Figure 6.
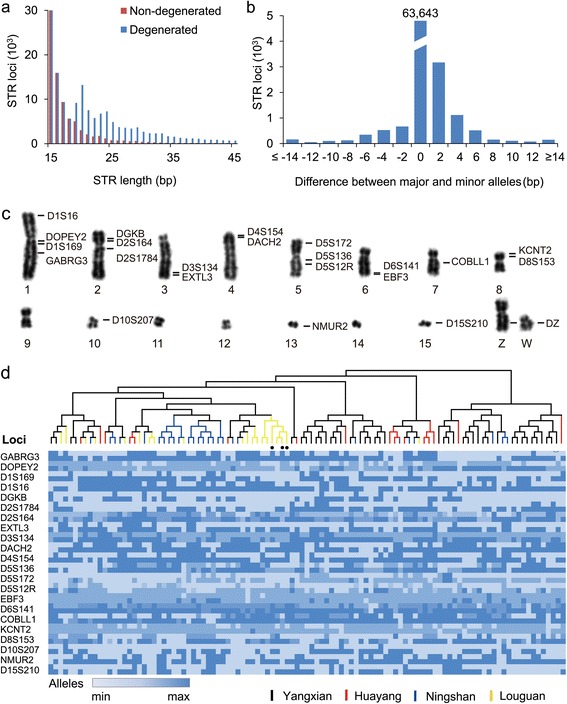
Genome-wide STR profiling of four ibis sub-populations. (a) STR (units of 2 bp, 3 bp, 4 bp, 5 bp, and 6 bp) distribution as a fraction of the total repeat length. Non-degenerate STRs do not contain insertions, deletions or mismatches. (b) Near random distribution of allele size differences between the major and minor alleles (n = 9). Size difference is calculated by subtracting the minor allele length from the major allele length. (c) Genetic markers of the ibis chromosomes typed in this study. Twenty-two representative STR and a single sex chromosome (W)-derived markers are shown here. (d) Individual identification based on the 22 STR loci. The colored horizontal scale bar indicates the number of repeat units (from minimum to maximum). The alleles (105 individuals) are used to construct neighbor-joining tree within sub-populations (Yangxian, n = 42; Ningshan, n = 27; Huayang, n = 16; and Louguan, n = 20). Solid circles denote the three individuals from a single family.
