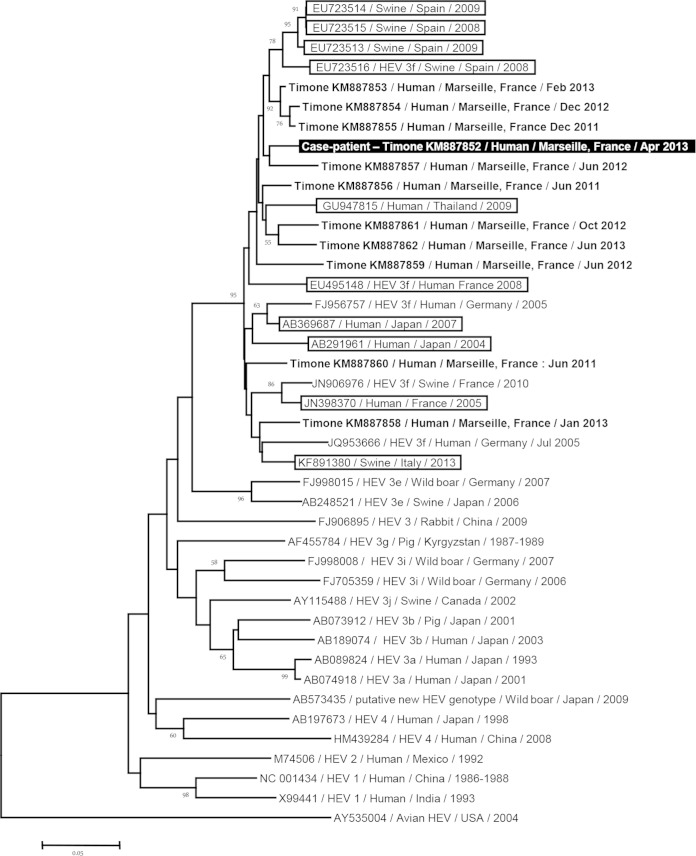
FIG 1.
Phylogenetic tree based on a 225-nucleotide partial sequence corresponding to nucleotides 6425 to 6649 of open reading frame 2 (ORF2) of the HEV genome (GenBank accession no. FJ956757). The HEV sequence obtained in the present study is indicated by a black background and a white font. The 10 sequences with the highest BLAST scores recovered from both GenBank (boxed) and Timone's database (boldfaced) were included, along with additional sequences representative of genotypes 1, 2, 3, and 4 and novel zoonotic strains. Nucleotide alignments were performed using the MUSCLE software (http://www.ebi.ac.uk/Tools/msa/muscle/). The tree was constructed using the MEGA 6 software (http://www.megasoftware.net/) and the neighbor-joining method. Branches with bootstrap values were obtained from 1,000 resamplings of the data, and values of >50% are labeled on the tree. The avian HEV sequence AY535004 was used as an outgroup. The scale bar indicates the number of nucleotide substitutions per site.