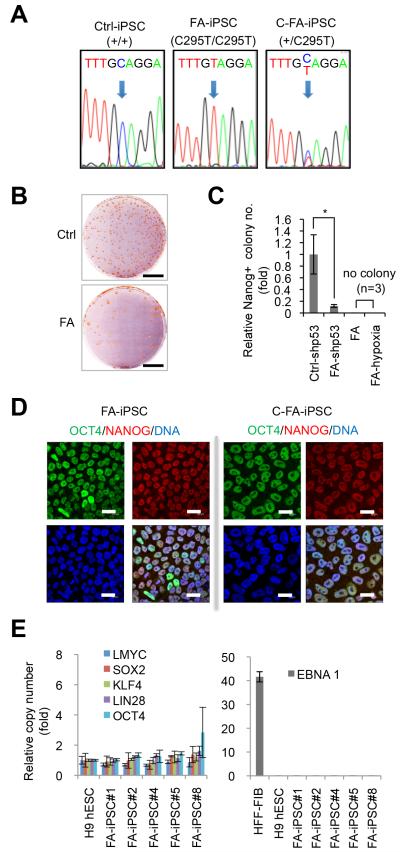
Fig. 1. Generation of FA-specific iPSCs.
A, DNA sequencing analysis revealed the presence of biallelic C295T point mutations in FANCA in FA-iPSCs, and the targeted correction of a FANCA-mutant allele in FA-iPSCs (C-FA-iPSCs). B, NANOG immunostaining of control (Ctrl) and patient (FA) colonies at day 25 and day 40 of reprogramming, respectively. Scale bar, 2 cm. C, Quantification of the number of NANOG-positive colonies at the end of reprogramming experiments. Numbers are normalized against control (mean±s.d., n=3, *p<0.05, t-test). shp53 indicates the use of p53 shRNA in the reprogramming cocktail. In both hypoxia (5%) and normoxia conditions, there were no NANOG-positive colonies without p53 shRNA. D, Immunofluorescence analysis of pluripotency markers OCT4 and NANOG in FA-iPSCs and C-FA-iPSCs. DNA was stained with Hoechst. Bar, 20 μm. E, Copy number quantification of reprogramming factor genes (left panel) and the episomal vector element EBNA1 (right panel). H9 human ESCs were included as a negative control. Human fibroblasts (hFib) 6 days after nucleofection were included as a positive control. The average copy numbers are comparable between H9 human ESCs and five randomly selected FA-iPSCs. Data are shown as mean±s.d. n=3.