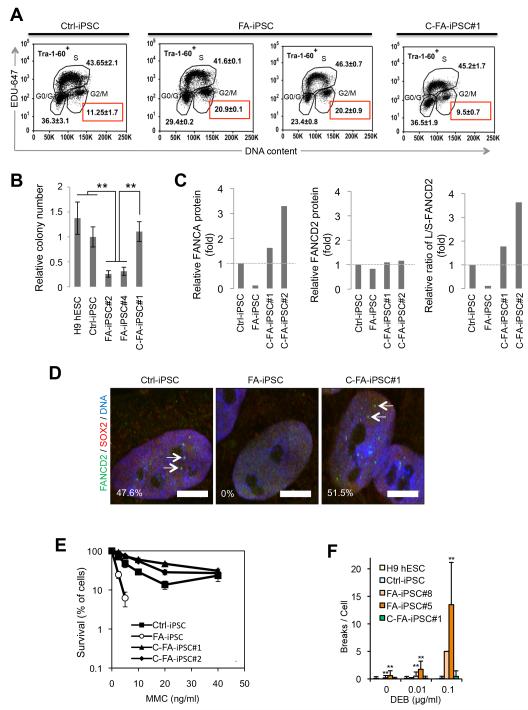
Fig. 3. FA-iPSCs recapitulate FA cellular defects.
A, FACS analysis of cell cycle profiles of the indicated iPSCs revealed an increased percentage of G2/M phase cells (indicated in red squares) in two randomly selected FA-iPSCs. C-FA-iPSC#1 indicates the targeted gene-correction clone. Values shown are mean±s.d. B, An identical number of iPSCs were seeded onto MEF feeder cells in the presence of ROCK inhibitor and allowed to form small colonies. The relative iPSC colony numbers were determined 10 days later. Data are shown as mean±s.d. n=3. **p<0.01 (t-test). C, MMC sensitivity of Ctrl-iPSCs, FA-iPSCs, C-FA-iPSCs#1, and FA-iPSCs lentivirally transduced with FANCA (C-FA-iPSC#2). Data are shown as mean±s.d. n=8. D, DEB induced chromosomal fragility test. Statistical analyses were performed by comparing Ctrl-iPSCs with other samples. Data are shown as mean±s.d. n=35 **p<0.01 (t-test). E, Western blotting analysis of FANCA and FANCD2 expression in indicated iPSC lines. WRN was included as a loading control. L-FANCD2 and S-FANCD2 indicate the mono-ubiquitinated and non-modified form of FANCD2, respectively. Quantitative analysis shows that targeted correction of the FANCA gene (C-FA-iPSC#1) or lentiviral introduction of FANCA in FA-iPSCs (C-FA-iPSC#2) restored expression of FANCA protein and mono-ubiquitination of FANCD2. F, Immunostaining of FANCD2 and SOX2 in the indicated iPSCs treated with 100 ng/ml MMC for 24 h. The percentage of nuclei positive for FANCD2 foci is indicated in the bottom left corner. Bar, 10 μm. Arrows denote FANCD2 foci.