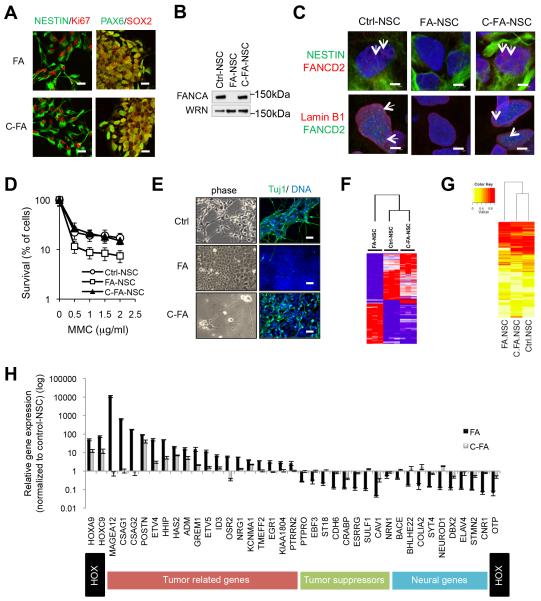
Fig 7. Cellular defects and molecular signatures of NSCs derived from FA-iPSCs.
A, Immunofluorescence analysis of neural progenitor markers in FA-iPSC derived NSCs (FA-NSCs) and C-FA-iPSC derived NSCs (C-FA-NSCs). Bar, 20 μm. B, Western blotting analysis of FANCA expression in control-iPSC derived NSCs (Ctrl-NSC), FA-NSCs and C-FA-NSCs. WRN expression was included as a loading control. Also see Supplementary Fig. 8. C, Immunostaining of FANCD2, lamin B1 and NESTIN in the indicated NSCs treated with 100 ng/ml MMC for 24 h. Arrows denote FANCD2 foci. Bar, 5 μm. D, MMC sensitivity of indicated NSCs. Data are shown as mean±s.d. n=8. E, Representative bright field (left panels) and Tuj1 immunofluorescence (right panels) micrographs of cultures spontaneously differentiated from Ctrl-, FA-, and C-FA-NSCs. DNA was counterstained with Hoechst. Bar, 50 μm. F, Hierarchical clustering analysis of genes with a minimum 3-fold difference in both comparisons (Ctrl-NSC vs. FA-NSC; FA-NSC vs. C-FA-NSC). 96% of genes (97 out of 101) altered by the FA mutation were rescued in gene corrected NSCs. Also see Supplementary Data 1. G, Heatmap and hierarchical clustering of DNA methylation levels at CpG sites in the promoter regions of the genes rescued by C-FA-NSC. Note that not every gene rescued by C-FA-NSC from gene expression analysis showed differential DNA methylation on their promoter regions. H, RT-qPCR analysis of the expression of selected genes in passage 2 NSCs derived from Ctrl-, FA-, and C-FA-iPSCs. The expression levels of genes in Ctrl-NSCs were set to one. Data are shown as mean±s.d. n=3. Gene functions are annotated below gene names.