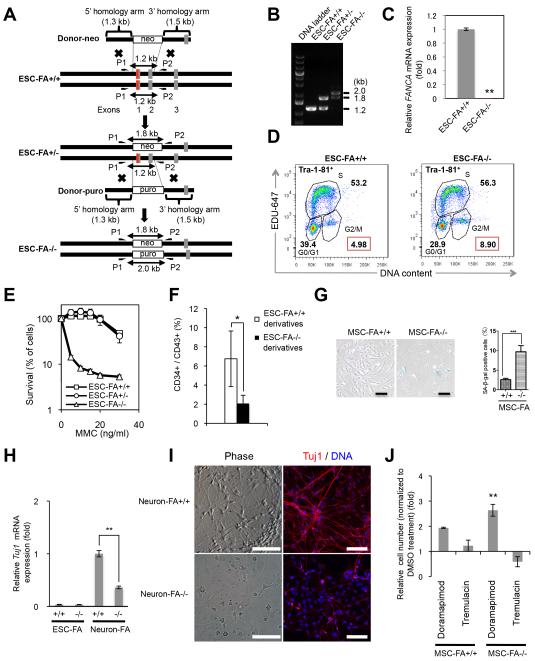
Fig. 9. Generation and characterization of FANCA knockout ESCs.
A, Schematic representation of TALEN-based knockout of the FANCA gene. The donor vectors include a neomycin-resistant cassette (neo) or a puromycin-resistant cassette (puro). Half arrows indicate primer sites for PCR (P1 and P2). The red line indicates the TALEN target site in exon 1. The human H9 cells were used as wild type host cells (ESC-FA+/+). The heterozygous FANCA mutant ESC line (ESC-FA+/−) was generated by one round of gene targeting, and the biallelic FANCA knockout mutant ESC line (ESC-FA−/−) was generated by a 2nd round of gene targeting. B, PCR analysis of ESC-FA+/+, ESC-FA+/− and ESC-FA−/− using P1 and P2 primer pairs shown in (A). M, DNA ladder. C, RT-PCR analysis of ESC-FA+/+ and ESC-FA−/−. ESC-FA−/− did not express FANCA mRNA. Data are shown as mean±s.d. n=3. ** p<0.01 (t-test). D, FACS analysis of cell cycle profiles of the indicated ESCs revealed an increased percentage of G2/M phase cells (indicated in red squares) in FANCA knockout cell (ESC-FA−/−). E, MMC sensitivity of ESC-FA+/+, ESC-FA+/− and ESC-FA−/−. Data are shown as mean±s.d. n=8. F, Percentage of differentiated ESCs that are CD34+/CD43+. Error bars represent SEM of 3 independent experiments. * p<0.05 (t-test). G, Representative SA-β-galactosidase staining (left panel, Scale bar, 200 μm) in passage 1 MSCs derived from ESC-FA+/+ and ESC-FA−/−, and quantitative data (right panel). H, RT-qPCR analysis of TUJ1 in ESCs and their pan neuron derivatives. Data are shown as mean±s.d. n=3. ** p<0.01 (t-test). I, Representative bright field (left panels) and Tuj1 immunofluorescence (right panels) micrographs of cultures spontaneously differentiated from NSC-FA+/+ and NSC-FA−/−. DNA was counterstained with Hoechst. Bar, 100 μm. J, Quantification of accumulated doubling population of FA-MSCs with doramapimod or tremulacin treatments at passage 5. The drug effects were normalized by the number of DMSO treated cells. Data are shown as mean±s.d. n=3. * p<0.05 (t-test).