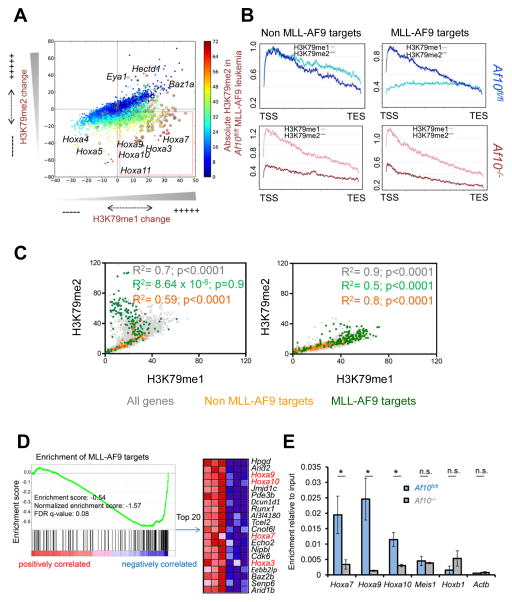
Figure 5. Genomic and epigenomic changes upon Af10 deletion.
(A) Scatter plot showing genome-wide changes in the values of promoter-proximal H3K79me1 (x-axis) or H3K79me2 (y-axis) in Af10fl/fl vs. Af10−/− leukemias. Each dot represents the difference in averaged methylation values around the promoter proximal regions (−2 kb to +2 kb around the TSS); positive and negative values reflect increases and decreases respectively in Af10−/− compared to Af10fl/fl leukemias. Methylation values are colored according to absolute H3K79me2 values in wild type MLL-AF9 leukemia (blue- least methylation, red-highest methylation). (B) A representative meta-analysis plot showing averaged profile across the gene body from the TSS to the transcription end site (TES) of MLL-AF9 target genes compared to control genes. Profiles of Af10fl/fl MLL-AF9 leukemias in (blue) compared to their Af10−/− counterparts (in red) are presented. Light colors represent H3K79me1 and dark colors represent H3K79me2. MLL-AF9 targets are genes bound by MLL-AF9 and non MLL-AF9 target genes are a size and expression matched set of genes that are not bound by MLL-AF9. (C) Scatter plot showing the genome-wide relative relationship between H3K79me1 and H3K79me2 values around promoter-proximal regions. Values are representative of 5 independent replicates for each group. (D) Enrichment of MLL-AF9 target genes in the Af10−/− compared to Af10fl/fl leukemias shown by GSEA. The top 20 MLL-AF9 targets downregulated in Af10−/− leukemia are shown on the right. (E) ChIP for DOT1L protein at the promoter proximal regions of indicated genes in MLL-AF9 leukemias. n=2, * p<0.05, n.s.= not significant.