Figure 1.
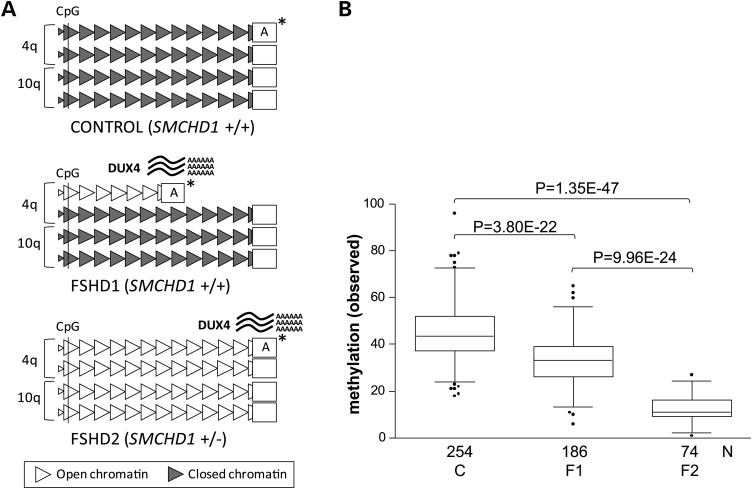
Overview of the genetic and epigenetic status at D4Z4 in control and FSHD. (A) Representation of the genetic and epigenetic status at the D4Z4 repeat arrays on chromosomes 4q and 10q in the control, FSHD1 and FSHD2 individuals. Normal-sized D4Z4 arrays (10–100 units) are depicted as 12 D4Z4 units (triangles), and the contracted array (1–10 units) in FSHD1 is depicted as 6 units. Gray-colored D4Z4 units represent a repressive chromatin state, which is partially lost at the contracted array in FSHD1 and at all four arrays in FSHD2 (white colored units). The chromatin relaxation is marked by a loss of CpG methylation at D4Z4, which is measured simultaneously on chromosomes 4 and 10 in the proximal D4Z4 unit (marked by CpG). The chromatin relaxation is associated with derepression of the DUX4 gene and stable DUX4 transcription from the permissive 4qA chromosomes (marked by asterisk). (B) Box plot showing the cumulative D4Z4 methylation level on chromosomes 4q and 10q D4Z4 arrays in 254 controls (C), 186 FSHD1 individuals (F1) and 74 carriers of an SMCHD1 mutation (F2) with significant differences between all groups (p values are indicated). The reduction in D4Z4 methylation in FSHD1 is caused by the presence of minimally one contracted repeat array. The box extends from the 25th to 75th percentiles, and the line in the middle of the box is plotted at the median, the whiskers are at the 2.5th and 97.5th percentiles.