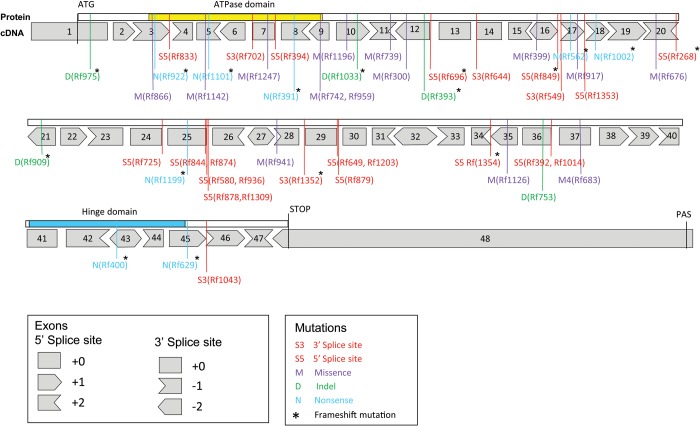
Figure 2.
SMCHD1 mutation spectrum in FSHD2. In 51 families carrying an SMCHD1 mutation, we identified 5 heterozygous insertion–deletion (indel) mutations (10%), 8 heterozygous nonsense mutations (15.5%), 14 heterozygous missense mutations (27.5%) and 24 heterozygous splice-site mutations (47%). Forty-five of the 51 SMCHD1 mutations were unique. In families Rf742 (from South of France) and Rf959 (from North of Spain), we identified the same missense mutation in exon 9. An identical splice-site mutation in exon 36 was found in two Dutch families (Rf392 and Rf1014). We found identical splice-site mutations in intron 29 in families from South Korea (Rf1203) and from the USA (Rf649). The highest incidence of shared mutations was found at the 5′ splice-site of intron 25, where three overlapping deletion mutations are identified in 6 families suggestive for a mutation hotspot (Supplementary Material, Fig. S2). All 48 SMCHD1 exons are indicated with boxes. The position of the 5′ and 3′ splice sites with respect to the ORF is also indicated and illustrated in the box below. The localization of the ATPase domain and the Hinge domain is indicated above the exon boxes. For all mutations, the nature of the mutation is indicated (M, D, N, S3 or S5, see box below) as well as the family number. An asterisk marks the mutations that disrupt the ORF.