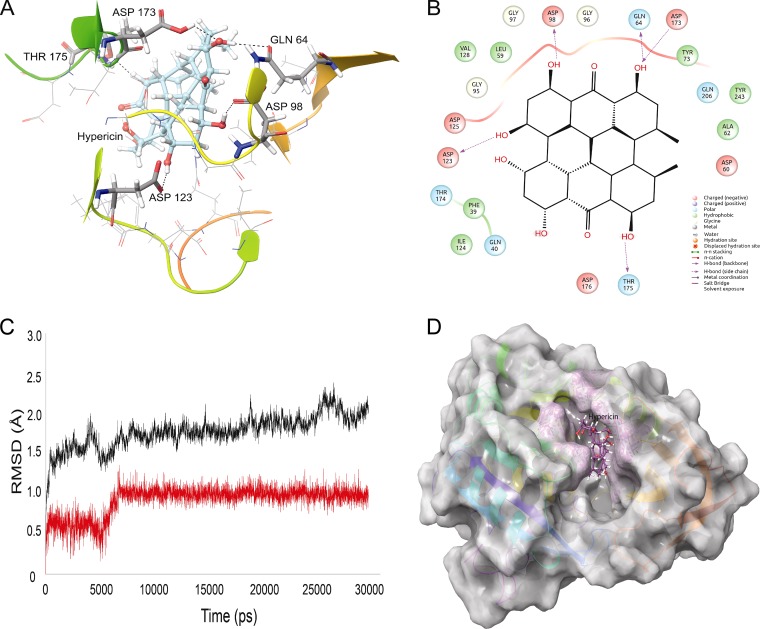
FIG 1.
Interactions of LdSS with hypericin in molecular docking simulations. (A) Hypericin interacting with the active site residues of LdSS. Hydrogen-bond-forming active site residues are displayed in tube representation and labeled. Asp173 and Thr175 belong to the gatekeeping loop of LdSS, which is a key structure in the opening and closing of the active site of LdSS. (B) Two-dimensional representation of interactions between LdSS and hypericin. (C) RMSD graph of the protein backbone (black) with hypericin at the active site (red). Clearly, overall the protein backbone is highly stable throughout the MD simulation, with very low RMSD values of ∼1.5 to 2.0 Å. Hypericin slightly adjusts its position within the active site of LdSS between 5 and 6 ns and becomes stable until the simulation is completed. (D) Hypericin (purple) buried in the active site of LdSS (white) after 30 ns of MD simulation.