FIG 1.
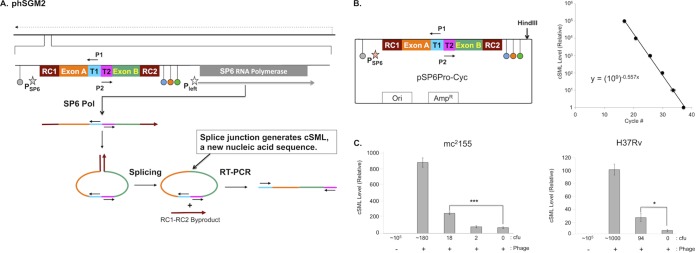
phSGM2 design, cSML detection assay dynamic range, and detection of M. smegmatis and M. tuberculosis. (A) The mycobacteriophage TM4 genome is depicted as a solid black line at the top of the figure. The direction of phage gene expression by host RNA polymerase is indicated by the dashed arrow above the phage genome. The site in the phage genome where the SGM is inserted is indicated and expanded. The SGM is comprised of two cassettes. The first is the SP6Pol ORF under transcriptional control of the mycobacteriophage L5 Pleft promoter (open star), which directs expression of SP6Pol during infection of mycobacteria. The SP6Pol ORF is codon optimized for efficient translation in M. tuberculosis, and translation initiation is directed by the Shine-Dalgarno sequence upstream of the TM4 major capsid subunit. The second cassette encodes the SP6 promoter (filled star) fused to a downstream sequence encoding several functional modules. The SP6 promoter cassette is flanked by transcription terminators. Upstream of the SP6 promoter is the E. coli rrnBT2 terminator (gray dot) (17), which precludes transcription of the SP6 promoter cassette by host RNA polymerase. We positioned three terminators downstream of the SP6 promoter cassette: phage SP6 major capsid subunit (SP6MCS; blue dot) (8), T7 (orange dot) (15), and the E. coli rrnC (green dot) (17) terminators. These terminators were designed to lower the frequency at which elongating SP6Pol transcribes the SP6Pol ORF. We also positioned several functional modules downstream of the SP6 promoter: T1 and T2 encoded sites to which primers P1 and P2 bind. These sites were oriented such that upon binding to T1 and T2, primers P1 and P2 are unable to generate a PCR amplification product. Adjacent to T1 and T2 were exons A and B. RC1 and RC2 were positioned at the 5′ of exon A and 3′ of exon B, respectively. RC1 and RC2 encode two halves of the RNA cyclase (RC) ribozyme (10). After transcription of this locus by SP6 Pol, a single-stranded RNA was synthesized that had one half of RC (RC1) fused to the 5′ end of exon A followed by T1, T2, and the other half of RC (RC2) fused to the 3′ end of exon B. Once RC2 was synthesized, RC1 and RC2 interacted and formed the active RC ribozyme, which mediated circularization of the single-stranded RNA between RC1 and RC2 by fusing exons A and B via a splicing reaction. Fusion of exons A and B created a new nucleic acid sequence distinct from the cognate DNA locus in the phage genome and constituted generation of the cSML. In addition, RC1 and RC2 fused to each other as a by-product. Finally, detection of the cSML was performed using P1 and P2 because the splicing of exons A and B generated an intervening sequence between the 3′ ends of P1 and P2, resulting in the creation of a template for RT-PCR amplification. The cSML was amplified by using a one-step, combined RT-PCR and detected by a molecular beacon. (B) The dynamic range was determined by cSML synthesis from HindIII-digested pSP6Pro-Cyc by in vitro transcription. Duplicate 10-fold serial dilutions were prepared, and cSML was amplified. The average cycle detection thresholds for each dilution were plotted, and the best-fit line through the data points is described by the equation y = (109)−0.557x, where x is the cycle detection threshold, and y is the relative level of cSML in a sample. This equation is used to determine the relative level of cSML present in a sample. (C) Determination of the lowest numbers of M. smegmatis mc2155 and M. tuberculosis H37Rv detected by the assay.
