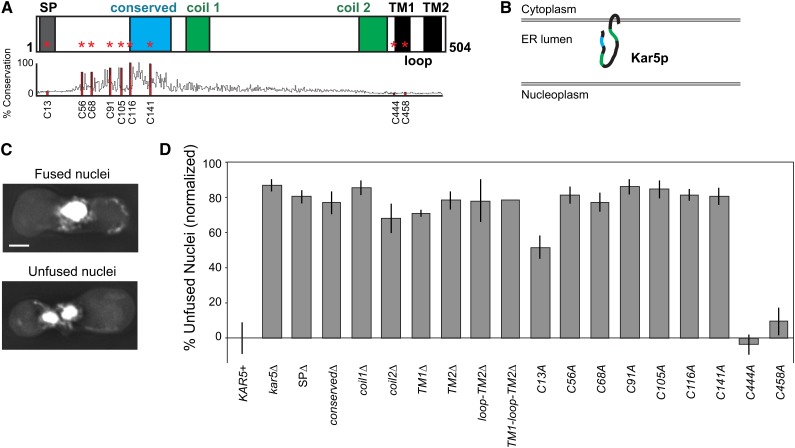
Figure 1.
All major Kar5p domains are essential for nuclear fusion. (A) Schematic of Kar5p predicted domains (Beh et al. 1997). Cysteines are marked with a red asterisk. Percent conservation is the percent identity to the consensus sequence among 96 Pfam sequences (Family Tht1, PF04163). SP refers to signal peptide (4−23). Coil1 and coil2 refer to two predicted coiled-coil domains (186−215 and 401−436). The conserved domain runs from residues 116 to 167. TM refers to transmembrane domains (TM1 445−465, TM2 481−504). The loop domain refers to the small cytoplasmic loop between the TM domains (466−480). Specific residues deleted in each construct are listed in the strain table (Table S1). (B) Schematic of Kar5p predicted topology based on protease protection assays and computational predictions (Beh et al. 1997). Note that SP is likely cleaved and thus not shown. The lumenal secondary structure is completely speculative. (C) Representative examples of fused and unfused nuclei. Scale bar, 2 μm. (D) Nuclear fusion assay. All crosses were kar5Δ (MS7670) × kar5Δ (MS7673). MS7670 contained a CEN plasmid with the indicated kar5 alleles. To remove the effects of plasmid loss, values were normalized against KAR5+ (29% unfused nuclei). Average of multiple independent experiments are shown (at least three trials for each, except two trials for SPΔ, conservedΔ, coil2Δ, TM1Δ, TM1-loop-TM2Δ, and C458A). Error bars show ± SEM.