Abstract
Transcription initiation by RNA Polymerase II (Pol II) is an essential step in gene expression and regulation in all organisms. Initiation requires a great number of factors, and defects in this process can be apparent in the form of altered transcription start site (TSS) selection in Saccharomyces cerevisiae (Baker’s yeast). It has been shown previously that TSS selection in S. cerevisiae is altered in Pol II catalytic mutants defective in a conserved active site feature known as the trigger loop. Pol II trigger loop mutants show growth phenotypes in vivo that correlate with biochemical defects in vitro and exhibit wide-ranging genetic interactions. We assessed how Pol II mutant growth phenotypes and TSS selection in vivo are modified by Pol II genetic interactors to estimate the relationship between altered TSS selection in vivo and organismal fitness of Pol II mutants. We examined whether the magnitude of TSS selection defects could be correlated with Pol II mutant-transcription factor double mutant phenotypes. We observed broad genetic interactions among Pol II trigger loop mutants and General Transcription Factor (GTF) alleles, with reduced-activity Pol II mutants especially sensitive to defects in TFIIB. However, Pol II mutant growth defects could be uncoupled from TSS selection defects in some Pol II allele-GTF allele double mutants, whereas a number of other Pol II genetic interactors did not influence ADH1 start site selection alone or in combination with Pol II mutants. Initiation defects are likely only partially responsible for Pol II allele growth phenotypes, with some Pol II genetic interactors able to exacerbate Pol II mutant growth defects while leaving initiation at a model TSS selection promoter unaffected.
Keywords: transcription initiation, GTFs, gene expression, transcription start site, RNA polymerase, trigger loop
Pol II is essential for expression of all protein-coding genes, and determining how the combined defects of Pol II activity mutants in all steps of the Pol II cycle (i.e., initiation, elongation, termination, cotranscriptional events) lead to growth defects is a difficult task. Initiation, the first step in transcription, is highly conserved, with regulation requiring a great number of factors (Hahn 2004; Cramer et al. 2008). Classical biochemical experiments using model promoters have shown that Pol II requires general transcription factor (GTF) TFIID, TFIIB, TFIIF, TFIIE, TFIIH for promoter recognition, formation of the preinitiation complex (PIC), promoter melting, and transcription start site (TSS) selection. The integration of these and other factors determines the efficiency of any particular promoter and the sequences that will be used to initiate transcription. Much remains to be understood about the functions of GTFs and how they integrate with Pol II activity during initiation. The effects of Pol II activity and roles of GTFs in initiation are especially visible in the process of TSS selection in S. cerevisiae, where most promoters use multiple start sites, and this usage is sensitive to a number of factors. Therefore, examination of mutant effects on TSS selection allows a window to the initiation process in vivo.
The most well-known core promoter element, the TATA box (consensus TATAWAWR motif in S. cerevisiae), is highly conserved throughout evolution but is only found at a subset of promoters. TSS selection at TATA element-dependent promoters in S. cerevisiae involves recognition of transcription start sites positioned 40–120 nucleotides (nt) downstream from the TATA box and the use of multiple start sites at most promoters, whether they are classified as TATA-containing (contains consensus TATA box) or not (Li et al. 1994; Dvir 2002; Basehoar et al. 2004; Corden 2008) (H. Jin and C. D. Kaplan, unpublished observations). Such extensive downstream positioning of TSSs in yeast is distinct from other eukaryotes for TATA-dependent promoters, where starts are more tightly focused ∼30 nt downstream of the beginning of the TATA box. Despite this difference in TSS distance to promoter element, in S. cerevisiae promoter melting appears to start 20–30 nt downstream of the TATA box and thus is similar to higher eukaryotes even though start sites can be more than 100 nt further downstream in S. cerevisiae (Giardina and Lis 1993). These results suggested that S. cerevisiae Pol II scans for favorable TSSs subsequent to promoter melting and open complex formation (reviewed in Kaplan 2013). A directional model for TSS scanning is strongly suggested from mutational analysis of start site regions and the distribution of TSSs when efficient start sites are compromised (Kuehner and Brow 2006; Kostrewa et al. 2009). The contribution of scanning to TSS usage in other organisms is unknown; however, the majority of promoters in higher eukaryotes utilize multiple, dispersed TSSs in a manner at least superficially analogous to S. cerevisiae (Choi et al. 2002; Hoskins et al. 2011; FANTOM Consortium et al. 2014; Haberle et al. 2014).
GTFs TFIIB (encoded by SUA7 in S. cerevisiae) and TFIIF (encoded by TFG1, TFG2, and TFG3 in S. cerevisiae) contribute to TSS selection. TFIIB bridges the TATA binding protein (TBP)-promoter DNA complex and Pol II, and likely stabilizes open complex formation through interacting with single-stranded DNA sequences in the PIC; TFIIF guides and stabilizes Pol II binding during assembly of PIC and appears to functionally interact with TFIIB, and may directly regulate Pol II activity (Henry et al. 1994; Sun and Hampsey 1995; Hampsey 1998; Chen and Hampsey 2004; Chen and Hahn 2004; Eichner et al. 2010; Luse 2012). A number of tfg1 and tfg2 alleles have been shown to shift distribution of TSSs toward upstream positions (Ghazy et al. 2004; Freire-Picos et al. 2005; Majovski et al. 2005; Khaperskyy et al. 2008; Eichner et al. 2010; Hahn and Young 2011). Conversely, mutations in SUA7 generally have been shown to alter TSS distribution toward downstream positions (Pinto et al. 1992, 1994; Hull et al. 1995; Sun and Hampsey 1995; Wu et al. 1999; Faitar et al. 2001; Chen and Hampsey 2004). Combination of TFIIB and TFIIF alleles can confer mutual suppression of their respective TSS defects along with TFIIF alleles’ suppression of TFIIB alleles’ temperature-sensitive phenotypes (Sun and Hampsey 1995; Ghazy et al. 2004; Freire-Picos et al. 2005). In addition, alleles of SSL2, which encodes an ATPase/helicase enzymatic subunit of TFIIH, have been shown to shift distribution of TSSs toward upstream slightly, and one allele that was shown to partially suppress downstream TSS shifts the cold sensitivity of sua7-1, suggesting functional importance of Ssl2 in TSS selection (Goel et al. 2012). Alleles of some Pol II subunits have been shown to affect TSS usage distribution on their own and that of GTF alleles when combined. Combination of alleles in tfg1 and rpo21/rpb1 resulted in suppressed temperature sensitivity and TSS defects of an rpo21/rpb1 allele (Freire-Picos et al. 2005). Combination of a tfg1 allele and rpb9∆ resulted in exacerbated TSS defects and temperature sensitivity (Ghazy et al. 2004). Alleles of rpb2 and rpb9 were shown to suppress the downstream shift effect and cold temperature sensitivity of certain sua7 alleles (Sun and Hampsey 1996; Sun et al. 1996; Chen and Hampsey 2004). Thus, for some GTF alleles, effects on TSS selection parallel their effects on growth phenotypes when combined with rpb alleles.
Our characterization of the relationship between Pol II catalytic activity mutants and TSS defects suggested an activity-based framework for interpretation of Pol II mutant TSS defects (Kaplan et al. 2012; Braberg et al. 2013). A mechanistic explanation for the connection of Pol II activity and TSS selection will be critical for understanding Pol II initiation. Mutations in Pol II subunit-encoding genes RPO21/RPB1, RPB2, RPB7, and RPB9 have been previously shown to alter TSS utilization in vivo, however, the mechanism of the alteration has been unclear (Hull et al. 1995; Sun and Hampsey 1996; Sun et al. 1996; Chen and Hampsey 2004; Freire-Picos et al. 2005; Chen et al. 2007; Kaplan et al. 2012; Braberg et al. 2013). In previous work, it was shown that rpo21/rpb1 mutants with substitutions in the trigger loop (TL), a mobile portion of the Pol II active center, have altered elongation rates in vitro. One class of these Pol II mutants confers faster elongation rates [termed gain of function (GOF)] in vitro, another class confers slower elongation rates [termed loss of function (LOF)], and the two classes are generally mutually suppressive when combined. These Pol II catalytic activity mutants conferred various phenotypes both in vitro and in vivo, including altered TSS selection, RNA splicing, presumed termination or processing defects, and chromosome segregation defects (Kaplan et al. 2008; Kireeva et al. 2008; Kaplan et al. 2012; Larson et al. 2012; Braberg et al. 2013; Viktorovskaya et al. 2013). The severity of TSS defects in vivo in both GOFs and LOFs correlated well with the extent of their deviation from WT elongation rate in vitro.
We previously found that Pol II GOF mutants shifted the distribution of TSSs upstream at ADH1 and other genes, similarly to tfg2 alleles and rpb9∆; conversely, Pol II LOF mutants shifted distribution of TSSs downstream at ADH1, similarly to most sua7 alleles (Kaplan et al. 2012; Braberg et al. 2013). Directional alteration of ADH1 TSS distribution by Pol II mutants, both GOFs and LOFs, mimic their effects on TSS distributions genome wide (H. Jin and C. D. Kaplan, unpublished results). Just as with the severity of their TSS defects, these Pol II mutants have growth defects in vivo that correlate with the extent of Pol II activity alteration. Mutants that have more severely altered activity in vitro (both fast and slow) show greater growth defects, more genetic interactions, and greater alterations to gene expression profiles in vivo. Growth defects can be suppressed when Pol II LOF and GOF mutations are combined within the same enzyme; similarly, there is mutual suppression of Pol II mutant TSS distribution defects at ADH1 in the double mutant, indicating a correlation between TSS defects and general growth defects (Kaplan et al. 2012). Because most Pol II mutant phenotypes we have studied correlate with strength of observed biochemical defects, it is difficult to distinguish whether observed in vivo growth defects derive especially from defects in a particular facet of transcription. Through genetic experiments and examination of TSS selection in vivo, we have attempted to understand further the relationship between Pol II activity defects, GTF function, and transcription initiation in S. cerevisiae. We also set out to extend our previous studies of factors that genetically interact with Pol II (genetic interactors) (Braberg et al. 2013) to understand their roles in TSS selection and the relationship between initiation and growth defects of Pol II alleles.
Materials and Methods
Yeast strains and media
Plasmids containing tfg2∆146-180, tfg2∆261-273, tfg2∆233-248 alleles were gifts from Steve Hahn (Eichner et al. 2010), plasmids containing sua7-1, sua7-3 were gifts from Michael Hampsey (Chen and Hampsey 2004). The sua7-58A5 and sua7-70A5 alleles were generated by Quickchange site-directed mutagenesis according to directions of the manufacturer (Stratagene/Agilent). Fragments containing target alleles were cloned into the yeast integrating vector pRS306 (Sikorski and Hieter 1989) and transformed into a strain background used for phenotyping and primer extension assay. For the complete list of yeast and bacterial strains used in this study, see Supporting Information, Table S1. Please see Supporting Information for note on the rpb1 mutant N1082S used in these studies.
Yeast media used in phenotyping assays were made as previously described (Amberg et al. 2005; Kaplan et al. 2012; Braberg et al. 2013). YP media contained yeast extract (1% w/v; BD), peptone (2% w/v; BD), and bacto agar (2% w/v; BD) supplemented with adenine and tryptophan. YPD media contained dextrose (2% w/v, VWR), YPRaf media contained raffinose (2% w/v, Amresco), and YPRafGal media contained raffinose (2% w/v) and galactose (1% w/v; Amresco) as the carbon source. YPRaf and YPRafGal media also contained antimycin A (1 µg/ml; Sigma-Aldrich). Synthetic complete media were made with “Hopkins mix” with certain amino acids dropped out at the concentrations described in Kaplan et al. (2012) after the slight modifications of Amberg et al. (2005). SC-Leu+MPA media contains 20 µg/ml final concentration of mycophenolic acid (Sigma-Aldrich) from a 10 mg/ml concentrated stock in ethanol (stored at −20°).
Mycophenolic acid (MPA) lowers cellular concentration of GTP by inhibition of IMPDH activity and induces expression of IMD2, which encodes an MPA-resistant form of IMPDH. Transcription mutants that are sensitive to lower GTP levels or those defective in induction of IMD2 confer MPA sensitivity (MPAs). For Pol II trigger loop mutants, MPA sensitivity is predictive of upstream start site defects at ADH1 (Braberg et al. 2013).
Strains used in this study contain the lys2-128∂ allele (Simchen et al. 1984) that renders cells auxotrophic for lysine due to a Ty1 retroelement long terminal repeat (LTR) insertion in the 5′ end of LYS2. Mutants that alter transcription at the allele can grow on medium lacking lysine (e.g., SC-Lys), a phenotype referred to as Spt− (Suppressor of Ty). The gal10∆56 allele (Greger et al. 2000; Kaplan et al. 2005) comprises a deletion in the GAL10 3′-UTR, resulting in compromised RNA processing and termination at GAL10, allowing transcription readthrough downstream into GAL7. Lack of GAL7 gene product allows accumulation of toxic intermediate products in galactose metabolism, thus WT gal10∆56 cells are sensitive to presence of galactose in the medium when GAL genes are expressed even in the presence of an additional usable carbon source (e.g., YPRafGal). Mutants that alter readthrough from gal10∆56 or otherwise increase GAL7 expression show galactose resistance on YPRafGal media, a phenotype referred to as GalR.
Primer extension assay for start site utilization detection
Primer extension assays were performed as previously described (Ranish and Hahn 1991) and with a few modifications described in (Kaplan et al. 2012). Briefly, 30 µg total RNA purified as previously described (Schmitt et al. 1990) was used to anneal with 32P-labeld oligonucleotide priming downstream of ADH1 start sites in 15 µl total reaction volume. Reverse-transcription reaction was performed by M-MLV reverse-transcriptase (Fermentas) in the presence of RNase Inhibitor (Fermentas) in 45 µl total reaction volume. Products were precipitated, digested with RNase A, and separated in 8% acrylamide gel made with 19:1 acrylamide:bisacrylamide (Bio-Rad), 1XTBE, and 7M urea, followed by visualization by phosphorimaging (Bio-Rad) and quantification with ImageQuant 5.1 software (GE).
Heatmaps for genetic interaction phenotypes
Growth on each media were scored using a 0–5 scoring system (0 = no growth, 5 = WT growth for all media except YPRafGal and SC-Lys; 0 = WT growth, 5 = growth of the mutant with maximum growth on the corresponding plate for YPRafGal and SC-Lys). To indicate growth difference in different growth conditions, all mutants on YPD, YPD 37°, YPRaf, SC-Leu phenotypes were normalized to WT on each plate by subtraction (mutant score−WT score). Negative numbers (slower growth) are shown as blue, positive numbers (faster growth) are shown in red, and inviable double mutants are in dark gray. Growth differences on SC-Leu+MPA were normalized to growth difference on SC-Leu by subtracting the difference on SC-Leu from the difference on SC-Leu+MPA, thus rendering net growth difference due to MPA sensitivity/resistance. Differences on YPRafGal and SC-Lys (WT growth is zero) were normalized to growth difference on YPD and SC-Leu (“standard growth condition” controls for these phenotyping media) by dividing the difference on YPRafGal or SC-Lys by ratio of WT growth to mutant growth on YPD or SC-Leu to quantify resistance phenotypes. Mutants that have GalR or Spt− phenotypes are thus shown as red in the heatmaps. Calculated score difference tables were turned into heatmaps using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html).
Results
Allele-specific genetic interactions between GTF mutants and Pol II trigger mutants
We used changes in model gene ADH1 TSS distribution as a proxy for in vivo initiation defects. To quantify changes in TSS distribution at ADH1, signals from ADH1 TSSs were placed into six bins and alterations in the fraction of TSSs present in each bin were determined relative to the WT distribution (Figure 1A). We first examined how GTF mutants—known to alter TSS on their own—altered TSS defects of Pol II mutants and whether they modulated Pol II mutant growth phenotypes to explore their possible influence on TSS defects of Pol II mutants and characterized any effects in light of any genetic interactions between Pol II mutants and GTF alleles. We wished to determine if opposite shifting Pol II and GTF TSS mutants were suppressive or additive when combined, for example, similarly to the combination of TFIIB and TFIIF alleles or combination of Pol II GOF and LOF mutants exhibiting suppression of TSS defects and growth phenotypes. Conversely, we might observe nonadditive behavior in double mutants, indicative of bypass or epistasis as we observed between sub1∆ and Pol II GOF alleles (Braberg et al. 2013). If a double mutant has a defect in growth phenotype that is better than expected from examination of individual phenotypes of single mutants [based on a multiplicative model for double mutant growth interactions (Schuldiner et al. 2006)] or an additive model for TSS defects, such an observation can be an example of epistasis. In such cases, single mutants would show a lack of independence when combined, with double mutants exhibiting phenotypes of one or the other single mutant, or a phenotype worse than either single mutant but to a lesser degree than would be expected from independently acting mutations. Finally, we asked whether GTF-Pol II genetic interactions and growth phenotypes strictly correlated with any observed modulation of Pol II mutant TSS defects.
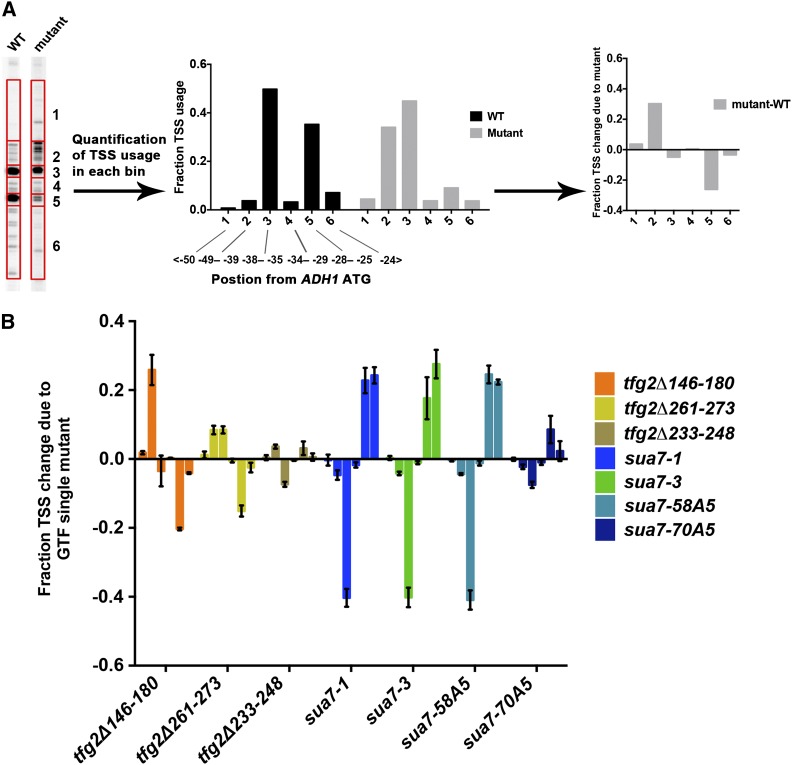
Figure 1.
Transcription start site (TSS) usage distribution at ADH1 and its alteration by Pol II GTF mutants. (A) TSSs detected by primer extension at ADH1 are distributed over a range of positions. To quantify TSS distributions, ADH1 start site signals were divided into six bins separated by promoter position and normalized to total signal for each lane (left panel). TSS usage distributions were quantified for different strains (middle panel). Relative change in normalized TSS usage distribution for mutant compared to WT (negative numbers indicate relative decrease in TSS position usage; positive numbers indicate relative increase) is then calculated and plotted (right panel). (B) Alterations in TSS usage at ADH1 caused by each GTF mutant shown were quantified as in (A). Start site defects of these GTF mutants are consistent with previous publications (Wu et al. 1999; Eichner et al. 2010), except for sua7-A5 alleles, which are in contrast to (Zhang et al. 2002). Graphs show average of at least three independent determinations with error bars representing SDs. See Figure S1 for representative primer extension experiments.
We integrated three deletion mutants of TFG2 into a yeast strain designed for phenotyping rpo21/rpb1 alleles. tfg2∆146-180 and tfg2∆261-273 had been shown to shift ADH1 TSSs upstream, whereas tfg2∆233-248 had been shown to exhibit a mild ADH1 TSS phenotype at best (Eichner et al. 2010). We also utilized SUA7 alleles containing substitutions in the “B-reader region”: sua7-1 (encodes E62K in TFIIB), sua7-3 (encodes R78C in TFIIB) alleles that have been shown to confer downstream TSS shifts by the Hampsey group (Pinto et al. 1994; Wu et al. 1999), and sua7-58A5 and sua7-70A5 mutants previously described as having upstream shift effects (Zhang et al. 2002). sua7-58A5 and sua7-70A5 each contain an insertion of five alanines at different positions in the B-reader (amino acid 58 or 70, respectively) and were recreated in our laboratory based on the published description by Zhang et al. (2002). Effects of GTF single mutants on ADH1 TSS distribution are shown in Figure 1B. tfg2∆146-180 has stronger upstream shifts at ADH1 than tfg2∆261-273, whereas tfg2∆233-248 has little effect, consistent with the work of Eichner et al. (2010). sua7-1 and sua7-3 show strong downstream shifts as shown previously (Pinto et al. 1994; Wu et al. 1999); however, sua7-58A5 and sua7-70A5 both show downstream shifts, strong and weak, respectively, which is typical behavior of sua7 TSS mutants but is opposite of published observations. We cannot explain this discrepancy but consider sua7-58A5 and sua7-70A5 to have standard behavior for sua7 alleles.
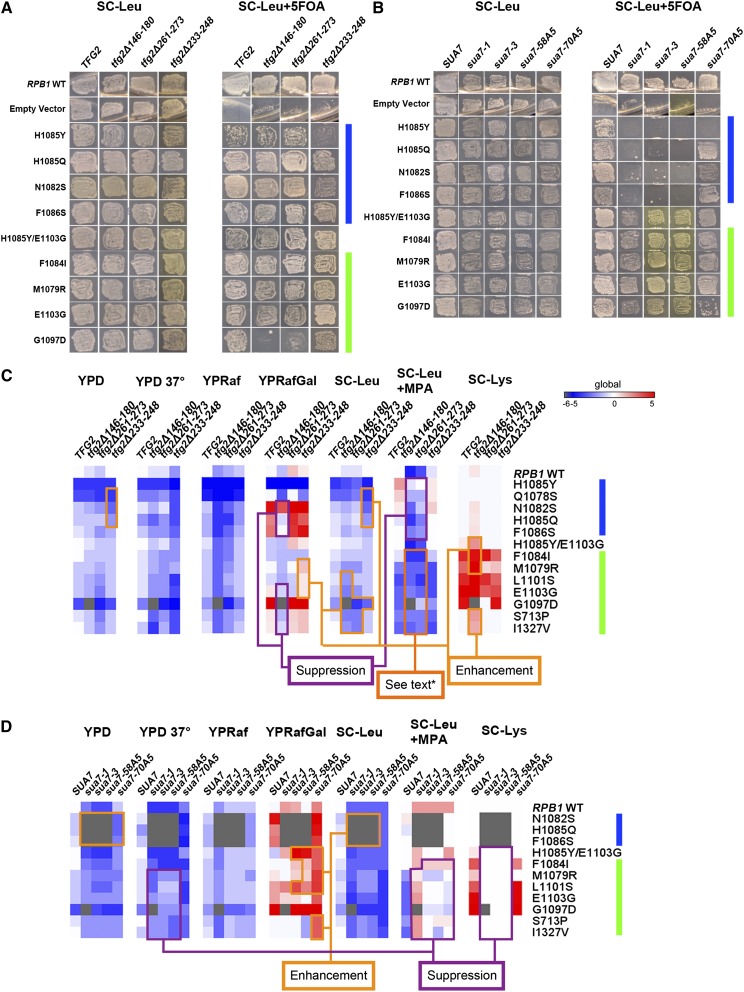
We tested the growth of these GTF mutants alone or in combination with Pol II mutants by transformation and plasmid shuffling of rpo21/rpb1 alleles in place of RPO21. Genetic interactions between GTF alleles and Pol II mutants appear complex, and we first describe genetic interactions apparent on growth of strains in rich or defined media (Figure 2, A and B; YPD and SC-Leu media described in Figure 2, C and D), followed by observed genetic interactions relating to gene-specific transcriptional phenotypes (Spt−, GalR, MPA sensitivity) (Figure 2, C and D).
Figure 2.
Genetic interactions between GTF and Pol II mutants. (A) Plasmids containing tfg2 mutants were integrated into yeast strains constructed to allow shuffling of a WT RPO21/RPB1 URA3 plasmid in favor of WT or mutant rpo21/rpb1 LEU2 plasmids through use of 5FOA poisoning of URA3+ cells. On the left, SC-Leu media allows coexistence of both URA3 and LEU2 plasmids. On the right, supplementation of SC-Leu with 5FOA uncovers rpb1 phenotypes by selecting against the RPB1 URA3 plasmid. Pol II mutants that have slower elongation rate than WT in vitro (LOFs) and mutants that genetically cluster with them are annotated with a blue bar; mutants that have faster elongation rate than WT in vitro (GOFs) and mutants genetically cluster with them are labeled with a green bar. Mutants are arranged by their measured elongation rates or elongation rates inferred by strength of genetic phenotypes compared with those mutants tested biochemically (Kaplan et al. 2012). (B) sua7 allele-Pol II mutant interactions examined as for tfg2 alleles in (A). (C and D) Phenotypes of viable GTF-Pol II double mutants are shown as a heatmap with qualitative determinations of growth defects on various media. Inviable double mutants are colored in gray. In YPD, YPD 37°C, YPRaf, and SC-Leu media, single and double mutant growth levels are normalized to WT. Blue indicates decreased growth relative to WT; red indicates increased growth compared with WT. In SC-Leu+MPA, growth difference is normalized to that on SC-Leu to quantify MPA sensitivity (shown as blue) or resistance (shown as red). In YPRafGal and SC-Lys, growth on the plate is divided by ratio of WT growth to mutant growth on corresponding general media (YPD and SC-Leu) to account for GalR and Spt- phenotypes (Gal+ and Lys+, shown in red) in contrast to their underlying growth defects. (C) tfg2 mutants. (D) sua7 mutants. See Figure S2 and Figure S3 for representative spot growth assay figures used for quantification in heatmaps and see Materials and Methods for further explanation of heatmaps.
When GTF alleles and Pol II mutants were combined, we observed allele-specific interactions between GTF alleles and Pol II GOF/LOF mutant classes. When Pol II GOF mutants and tfg2 alleles were combined, the most severe GOF mutant rpo21/rpb1 G1097D exhibited a strong negative interaction with tfg2∆146-180 or tfg2∆261-273 (lethality and synthetic sickness, respectively). LOF Pol II mutants (downstream TSS shifting mutants) and tfg2∆146-180 and tfg2∆261-273 (upstream TSS shifting mutants) did not result in suppression of growth defects of Pol II LOF mutants on standard rich or defined media in contrast to mutual suppression of growth defects when Pol II GOF and LOF (opposite TSS shifting mutants) are combined (Kaplan et al. 2012), or when tfg and sua7 alleles (opposite TSS shifting mutants) have been combined (Sun and Hampsey 1995; Ghazy et al. 2004; Freire-Picos et al. 2005) (Figure 2, A and C). The tfg2∆233-248 allele, which does not have a clear TSS defect, exhibited a negative interaction with Pol II LOF alleles but no clear interactions with Pol II GOF alleles. Neither tfg2 upstream shifting allele could rescue lethal LOF Pol II mutants (Figure S2A), in contrast to rescue of lethal Pol II LOFs when combined with GOF mutants (Kaplan et al. 2012).
When sua7 mutants were combined with GOF Pol II mutants (upstream TSS shifting mutants), we observed apparent lack of additivity in growth defects of strong downstream shifting sua7 alleles combined with Pol II GOFs. The double mutants generally showed growth defects in between those of the single mutants, suggesting directional suppression or epistasis, but not mutual suppression, where double mutants would be expected to grow better than either single mutant (mutual suppression is generally observed when Pol II LOF and GOF trigger loop mutants combined within the same enzyme). These effects were more pronounced on YPD medium than on YPRaf or defined medium (SC-Leu). We also observed partial suppression of the sensitivity to increased temperature (37°, Ts− phenotype) of sua7-1, sua7-3, and sua7-58A5 alleles by Pol II GOF alleles (Figure 2, Figure S3). The weak downstream shifting allele sua7-70A5 enhanced growth defects with Pol II GOF alleles, distinct from the other stronger sua7 alleles tested.
In contrast to the milder phenotypes of sua7-Pol II GOF strains, Pol II LOF mutants were exquisitely sensitive to defects in TFIIB, as widespread synthetic lethality or sickness was observed between sua7 alleles and Pol II LOF mutants. sua7-1, sua7-3, and sua7-58A5 showed very strong negative interactions with all LOF Pol II mutants tested, with most double mutants being inviable. A weak downstream shifting allele, sua7-70A5, showed negative but weaker interactions with Pol II LOF alleles. Both classes of mutant, Pol II LOF and sua7 alleles, alter TSS distributions in a similar fashion, suggesting exacerbated TSS defects might underlie observed synthetic genetic interactions (see below). These results indicate that combinations of Pol II and GTF mutants exhibiting the same polarity of TSS defects can lead to exacerbation of growth defects, wherein aggravated initiation defects may be a major contributor of the observed growth defects. The strongest genetic interaction between Pol II mutants and GTF alleles was observed when Pol II LOF mutants and strong sua7 downstream shifting alleles were combined (lethality), suggesting that Pol II LOF mutants are much more sensitive to initiation defects than GOF mutants. Combinations of mutants with opposing TSS distribution defects resulted in partial, but not necessarily mutual, suppression of single mutant growth defects on generic media.
We also examined conditional growth phenotypes on several other types of media, including those reporting on gene-specific transcription defects in vivo (Kaplan et al. 2012; Braberg et al. 2013). Results are shown as a heatmap of normalized estimates of phenotypic strength as determined by visual determination of growth on plates (tfg2 upstream shifting alleles in Figure 2C, sua7 downstream shifting alleles in Figure 2D; see Figure S2 and Figure S3 for representative images; see Materials and Methods for calculations). We observed sensitivity to mycophenolic acid (MPA) for tfg2 alleles, likely corresponding to upstream TSS shifts causing inability to induce IMD2 in the presence of MPA, with tfg2∆146-180 the most MPA-sensitive. MPA sensitivity has been shown to correlate well with upstream TSS shifts of a subset of Pol II mutants, including those tested here (Braberg et al. 2013). Conversely, sua7 alleles appeared resistant to MPA, similar to Pol II LOF alleles (Figure 2, C and D). When Pol II GOF alleles are combined with the strong upstream shifting allele tfg2∆146-180, the double mutants are inviable on this medium. The weaker upstream shifting allele tfg2∆261-273 also exacerbates MPA sensitivity of Pol II GOF alleles, with the resulting double mutants exhibiting no growth on this medium. This enhancement of MPA sensitivity leading to double mutants’ lack of detectable growth is not well-illustrated in our heatmap due to our calculation metric being unable to capture zero growth during calculation of “net” MPA sensitivity (see Materials and Methods). In contrast, MPA sensitivities of tfg2 alleles were suppressed when combined with Pol II LOFs; similarly, MPA sensitivities of Pol II GOFs were suppressed when combined with all sua7 alleles, being more apparent in stronger sua7 downstream shifting alleles. Therefore, MPA phenotypes of combinations of GTF alleles and Pol II mutants appeared additive or suppressive depending on the nature of the allele class: when upstream shifting alleles were combined, MPA sensitivity was exacerbated; when an upstream shifting mutant was combined with a downstream shifting mutant, MPA sensitivity was alleviated (Figure 2, C and D). These results support MPA sensitivity as a readout for initiation defects and not necessarily elongation defects, as widely assumed, and predict that TSS defects of Pol II mutants and GTF alleles may be additive and suppressive. sua7 alleles with strong TSS defects showed strong temperature sensitivity (Ts−); however, this could partially be alleviated when combined with Pol II GOF mutants (Figure 2D).
We also examined gene-specific transcription-related GalR and Spt− phenotypes, which have less clear relationships to TSS defects (see Materials and Methods). Most Pol II GOF mutants do show Spt− phenotype as measured by suppression of lysine auxotrophy (Lys−) in the presence of lys2-128∂ allele (see Materials and Methods), but there is only partial correlation with upstream shifting TSS defects. tfg2∆146-180 enhanced Spt− phenotypes of all moderate Pol II alleles, whereas the other tfg2 alleles did not show apparent interaction with Pol II alleles for the Spt− phenotype. Stronger downstream shifting sua7 alleles suppressed Spt− phenotypes of Pol II GOF alleles (Figure 2, C and D). A number Pol II LOF and GOF mutants have been shown to exhibit the GalR phenotype in the presence of the gal10∆56 allele of GAL10 (see Materials and Methods for description) (Kaplan et al. 2012). The strongest upstream shifting allele tfg2∆146-180 suppressed GalR phenotypes of all Pol II alleles that had the GalR phenotype, whether GOFs or LOFs; tfg2∆233-248, an allele with no apparent ADH1 TSS defect, enhanced GalR phenotypes of weak Pol II GOF alleles. sua7 alleles showed GalR phenotypes on their own and enhanced those of Pol II GOF alleles (Figure 2, C and D). The wide range of genetic interactions including enhancement and suppression of these conditional growth phenotypes suggests a complex network between Pol II and TFIIB/TFIIF that may relate to gene-specific effects not apparent in overall double mutant growth phenotypes.
Combinations of GTF alleles and Pol II alleles lead to mutual suppression of TSS defects but not mutual suppression of generic growth defects
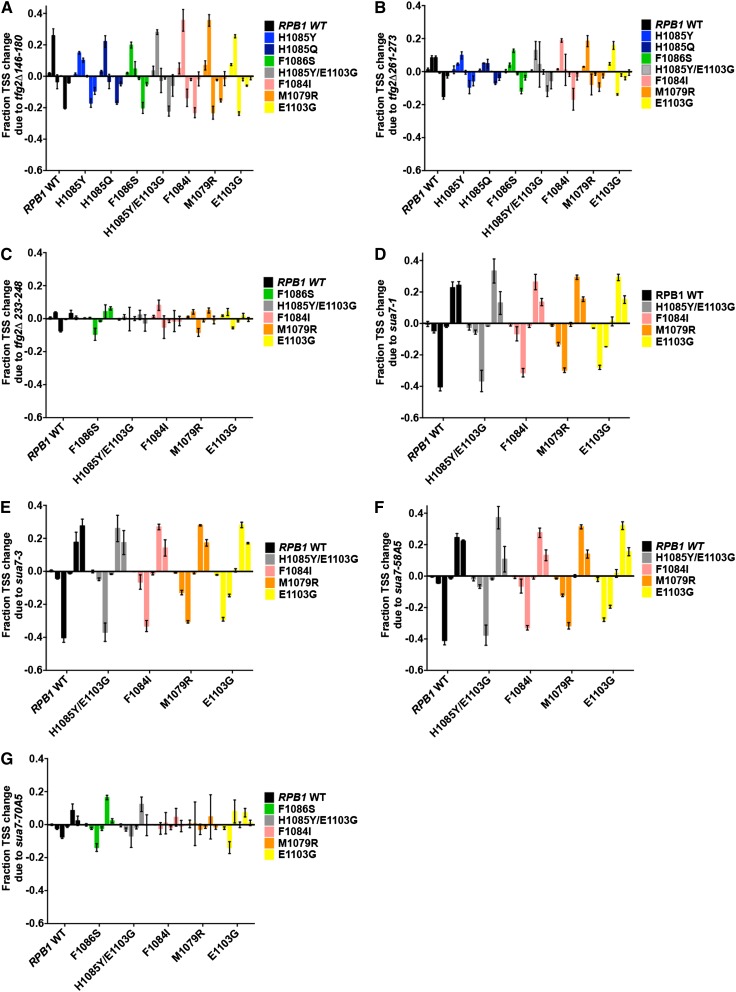
Because we observed above that GTF TSS defective mutants and Pol II TSS defective mutants conferred enhanced growth defects when same-direction TSS shifting mutants were combined but showed mostly weak, conditional, or directional suppression when opposite direction shifting mutants were combined, we examined how each class of double mutant affected TSS distribution at ADH1. This was performed to determine if TSS defects of combination of GTF alleles and Pol II mutants were additive and suppressive similar to MPA sensitivity phenotypes or were exacerbating, but not mutually suppressive, similar to general growth phenotypes of double mutants.
We observed that the TSS defects of GTF mutants and Pol II mutants were uniformly additive or suppressive, depending on the direction of TSS shifts of individual mutants, but not epistatic (Figure 3, see Figure S1 for representative raw data). tfg2∆146-180 and tfg2∆261-273 mutants shifted ADH1 TSSs upstream relative to all single Pol II alleles tested, indicating exacerbation of Pol II GOF alleles that shift ADH1 TSSs upstream on their own and suppression of Pol II LOF alleles that shift TSSs downstream on their own (Figure 3, A and B). The mutual suppression of TSS defects observed was similar to those observed for double mutant combinations of TFIIF and TFIIB alleles or for intra-Pol II double mutants. Conversely, those cases were accompanied by mutual suppression of growth phenotypes, which appears lacking for combinations of GTF alleles with Pol II trigger loop alleles (Figure 2). Additionally, strong downstream shifting alleles sua7-1, sua7-3, sua7-58A5 shifted ADH1 TSSs of sua7-Pol II GOF double mutants downstream relative to all Pol II GOF single mutants (Figure 3, D–F), indicating additive effects of opposite polarity shifts and, therefore, suppression of GOF Pol II allele TSS defects at ADH1. sua7-70A5, a weak downstream shifting allele, shifted ADH1 TSSs of sua7-70A5-Pol II double mutants downstream relative to all Pol II allele backgrounds (Figure 3G), indicating exacerbated downstream TSS shifts of LOF Pol II alleles and suppression of upstream TSS shifts of GOF Pol II alleles. TSS defects and generic and conditional growth phenotypes tested in GTF-Pol II double mutants suggest that TSS defects may contribute to general growth defects when TSS defects are severe, yet suppression of TSS defects (as measured at ADH1) does not correlate with suppression of Pol II mutant generic growth defects. Taken together, these results suggest that TSS defects may contribute to Pol II mutant growth defects, and Pol II activity alterations can partially compensate for defects in GTFs, but initiation defects are not likely to be the main or only drivers of observed Pol II allele growth phenotypes.
Figure 3.
Modulation of Pol II mutant TSS selection defects at ADH1 by GTF mutants. (A) Quantification of effects of GTF alleles on Pol II alleles on TSS utilization by comparison of double mutants to respective Pol II single mutants at ADH1 (quantified as in Figure 1A). Values indicate average of a minimum of three independent determinations, with SDs represented by error bars. (A.) tfg2∆146-180. (B) tfg2∆261-273. (C) tfg2∆233-248. (D) sua7-1. (E) sua7-3. (F) sua7-58A5. (G) sua7-70A5. See Figure S1 for representative primer extension experiments.
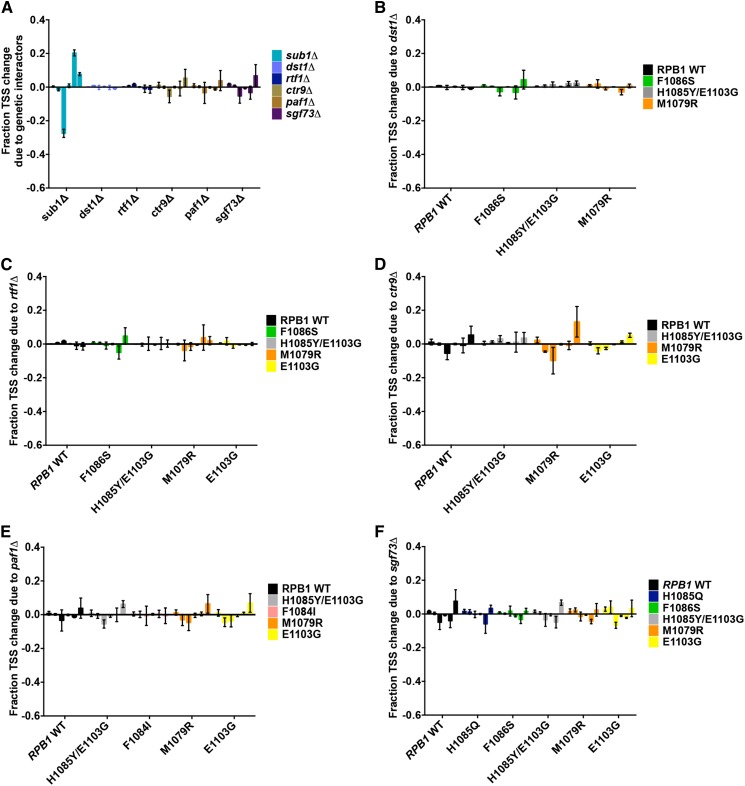
Genetic interactors with widespread genetic interactions with Pol II TSS defective alleles do not generally have TSS defects on their own or when combined with Pol II alleles
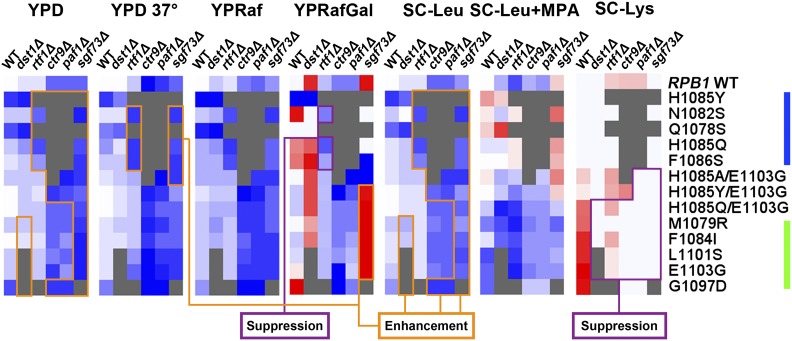
Inspired by the discovery of a Pol II genetic interactor, sub1∆, that conferred a downstream TSS defect on its own and enhanced downstream TSS defects and growth defects of LOF Pol II alleles but showed epistasis with GOF Pol II alleles for TSS defects and growth phenotypes (Braberg et al. 2013), we investigated how factors that have genetic interactions with Pol II TSS defective alleles affected TSS distributions on their own or in combination with Pol II alleles. Are genetic interactions of factors and Pol II TSS shifting alleles predictive of their effects on TSS utilization? The genetic interactions of dst1∆, rtf1∆, sgf73∆, paf1∆, and ctr9∆ with Pol II alleles have been shown previously (Hartzog et al. 1998; Malagon et al. 2006; Braberg et al. 2013). These and additional factors are illustrated in Figure 4. Deletion of DST1 (encoding the general transcription elongation factor TFIIS) showed allele-specific genetic interactions with GOF Pol II alleles and suppressed the Spt− phenotypes of Pol II alleles. RTF1, CTR9, PAF1 (genes encoding subunits of the Paf1C complex) showed stronger genetic interactions with Pol II LOF alleles and exhibited mild Spt− phenotypes on their own but suppressed the Spt− phenotypes of Pol II alleles. However, SGF73 (encoding a subunit of the histone-modifying SAGA complex) showed genetic interactions with both classes of alleles, enhanced MPA resistance of Pol II LOF alleles, and suppressed Spt− phenotypes of Pol II alleles (Figure 4). We found that deletions of these genetic interactors did not confer any strong TSS defects at ADH1 on their own (Figure 5A), nor did they modulate TSS defects of either LOF or GOF Pol II alleles (Figure 5B). The genetic interactions these factors exhibit with Pol II TSS-defective alleles may go through distinct mechanisms from sub1∆, whose genetic interactions on growth mirrored its effects on TSS defects at ADH1 when combined with Pol II alleles (Braberg et al. 2013). These results suggest genetic interactions between factors and Pol II alleles are not predictive of initiation defects, and that growth defects of many or most double mutant combinations with Pol II alleles do not result from exacerbation of TSS defects, under the assumption that ADH1 is a proxy for global TSS defects (supported by our unpublished global analysis of TSS defects in Pol II mutants).
Figure 4.
Genetic interactions between Pol II alleles and genetic interactor deletions. Phenotypes of genetic interactor deletions on their own or in combination with Pol II mutants on different medium normalized to WT are shown in the heatmap. See description in Figure 2, C and D and see Materials and Methods for heatmap details. See Figure S7B in Braberg et al. (2013) and Figure S5 for representative spot growth assays used for heatmaps.
Figure 5.
Genetic interactors do not generally modulate TSS defects of Pol II mutants at ADH1. (A) Quantification of TSSs usage alterations relative to WT at ADH1 in genetic interactor deletions are shown (quantified as in Figure 1A). Values indicate a minimum of three independent determinations, with SDs represented by error bars. (B–F) Quantification of TSSs usage distribution differences between genetic interactor deletion—Pol II double mutants relative to Pol II single mutants are shown. (B) dst1∆. (C) rtf1∆. (D) ctr9∆. (E) paf1∆. (F) sgf73∆, respectively. See Figure S4 for representative primer extension experiments.
Discussion
Combination of mutant alleles allows the relationships of different factors to be probed, with the hope of revealing distinctions between their contributions to different processes and possibly suggesting mechanism. A number of factors contribute to TSS selection in S. cerevisiae and their relationships have been probed here to understand the requirements for normal Pol II initiation. Previous analyses showing that distinct classes of TSS mutant exist in yeast (upstream and downstream shifting), supporting a scanning model for identification of TSSs. When mutants of differing TSS shift class have been combined (Sun and Hampsey 1995; Ghazy et al. 2004; Freire-Picos et al. 2005; Kaplan et al. 2012), they generally exhibited mutual suppression of TSS defects coupled to suppression of growth defects, whereas mutants of the same class exhibited enhancement of TSS and growth defects.
We previously discovered that SUB1 (homolog of PC4) has wide-ranging genetic interactions with Pol II mutants in a manner correlating with class of Pol II mutant (GOF or LOF) (Braberg et al. 2013). SUB1 was originally genetically isolated as a high copy suppressor of TFIIB alleles and biochemically as a positive transcription factor that stimulates basal transcription (Henry et al. 1996; Knaus et al. 1996). Deletion of SUB1 causes synthetic lethality in combination with sua7 TSS defective mutants (Knaus et al. 1996). The strong genetic interactions between SUB1 and SUA7 suggested a close association of their function in initiation and TSS selection and in vivo growth. We found that sub1∆ caused ADH1 TSS distribution to shift downstream and exacerbated the downstream shifts of Pol II LOF mutants, correlating with exacerbation of growth defects in sub1∆-Pol II LOF mutant double mutants (Braberg et al., 2013). Distinct from the general trend conferred by combination of TSS shifting alleles mentioned above, sub1∆ TSS effects were not additive with Pol II GOF mutants; instead, epistasis was observed for Pol II GOF alleles combined with sub1∆ (Braberg et al. 2013). This epistasis was in contrast to sub1∆ enhancement of Pol II LOF alleles for both growth phenotypes and TSS shifts. In light of these different classes of relationships previously observed among mutants altering TSS selection, we examined the relationships between GTF alleles and Pol II alleles with altered trigger loops and relationships between these same Pol II TL alleles and other Pol II genetic interactors.
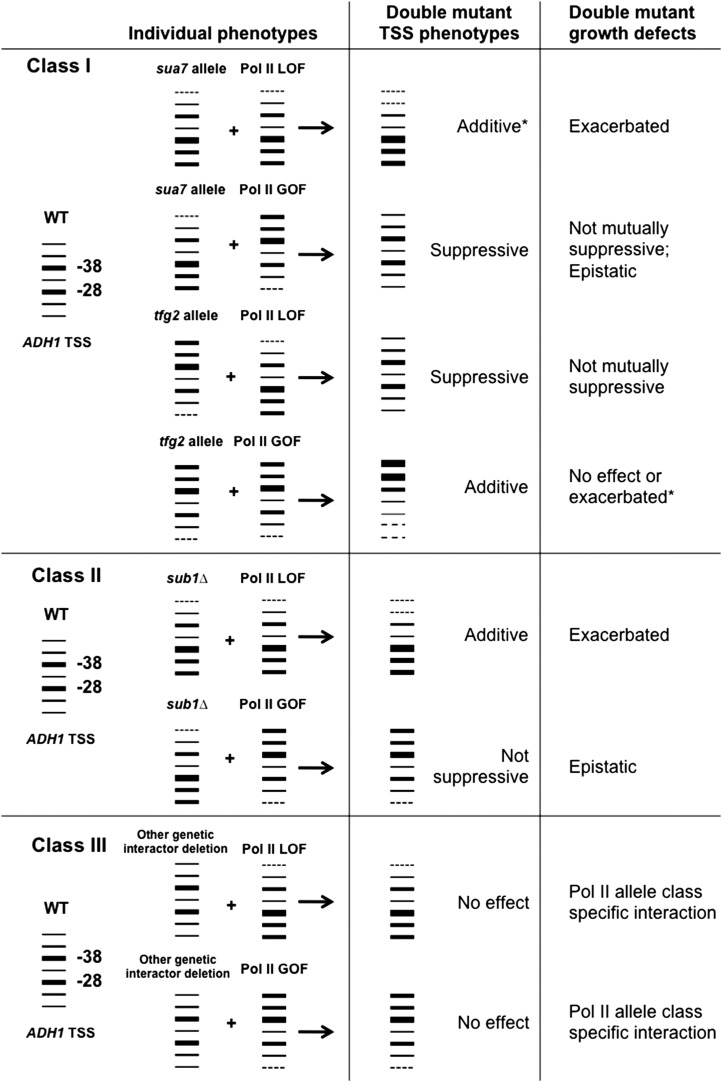
We found that, unlike previous combinations of TSS shifting alleles, suppression of TSS defects by Pol II mutant-GTF combinations could be partially separated from their effects on growth. Furthermore, we found that previous observations regarding the relationship of sub1∆ with Pol II alleles for TSS determination were relatively unique compared with a number of other Pol II genetic interactors examined here. Taken together, we can now discern at least three types of genetic relationships among TSS-altering alleles, Pol II interacting alleles, and Pol II active site mutants in regard to double mutant modulation of TSS and growth defects (Figure 6). First, GTF and Pol II mutants, each altering start sites on their own, have additive or suppressive effects on TSS distribution at ADH1, depending on the nature of their single mutant defects (Class I). Unlike combinations of GTF alleles and a subset of previously examined rpb alleles, TSS defect suppression is partially uncoupled from growth defect suppression for oppositely acting TSS alleles. Negative growth interactions between GTF alleles and Pol II alleles correlated with exacerbated TSS defects but were of much greater strength for downstream shifting GTF TSS alleles and Pol II LOF alleles than for upstream shifting GTF TSS alleles and Pol II GOF alleles. These observations suggest that S. cerevisiae growth is much more sensitive to defects in initiation that result from decreased initiation efficiency. Second, sub1∆ is thus far unique in the strong epistasis observed between Pol II GOF alleles and sub1∆ for TSS selection at ADH1 (Class II). This indicates that sub1∆ defects are distinct from those of TFIIB alleles, and that while sub1∆ appears to be bypassed in Pol II GOF alleles, sua7 defects are not, although there is some observable suppression of specific phenotypes in sua7-Pol II GOF mutant strains. Third, other tested genetic interactors with Pol II do not generally have TSS defects on their own or modulate Pol II TL mutant TSS defects (Class III), suggesting that the strong correlation between TSS defects and a very broadly Pol II interacting mutant, sub1∆, is relatively unique. Genetic interactions we observe between these factors and Pol II mutants may be originated (caused) by other transcriptional defects.
Figure 6.
Model of relationships between TSS distribution shifting GTF alleles, Pol II active site alleles, and other genetic interactors. GTF alleles and Pol II mutants have additive or suppressive effects on TSSs at ADH1 while showing exacerbation when same direction TSS mutants are combined, but no mutual suppression of growth defects when opposite polarity TSS mutants are combined (Class I); sub1∆ has enhancement or epistasis with Pol II alleles on both TSS defects and growth defects, which is thus far unique among tested mutants (Class II), whereas other tested genetic interactors do not modulate TSS defects on their own or in combination with Pol II alleles but exhibit a wide range of genetic interactions with Pol II TSS shifting mutants, suggesting relationships based on defects outside of TSS selection or initiation (Class III). *Description based on viable double mutants.
Although our experiments suggest that Pol II activity–dependent growth defects can be uncoupled from observed TSS defects, open questions remain concerning the mechanisms by which Pol II start sites are determined. RNA polymerases prefer to initiate at YR (−1, +1) sites, where the initiating nucleotide of an RNA is a purine, with a pyrimidine just upstream. A crystal structure of a viral RNA polymerase suggested that this sequence preference was likely due to purine stacking between the initiating NTP and a purine at the −1 position on the template strand (meaning a pyrimidine at −1 on the transcribed strand), and a set of very recent bacterial RNAP structures confirm this for multisubunit RNAPs (Gleghorn et al. 2011; Basu et al. 2014; Zhang et al. 2014). It seems likely that there are additional sequence determinants controlling efficiency of usage of any particular start site. For example, at ADH1 transcription mutants appear to alter the probability of usage of YR sequences that are used by WT present in the start region. Although Pol II GTFs might be positioned within the Pol II PIC to interact with sequences and “read” for the start site, the primary determinants for TSS preference are the −1/+1 bases that are located deep in the active site and not bases juxtaposed to hypothetical or predicted GTF locations in the PIC (these would mostly be upstream sequences). Moreover, the types of TSS changes observed for GTF mutants are phenocopied by mutations in the Pol II trigger loop, suggesting they might arise from similar types of defects in transcription. What could these similar defects be?
The altered patterns of starts observed in most upstream or downstream shifting start site mutants appear to be stereotypical to each class, meaning the positions of starts that are more likely used at ADH1 in mutants are the same within each mutant class as defined by upstream or downstream shifting, not by whether they are in GTF subunits or the Pol II active site. However, the fraction of usage on these usable start sites differs (strong or weak shift in distribution of TSS usage) depending on how much an allele deviates from WT catalytic activity for Pol II alleles or on how strong they appear genetically (for GTF alleles). In other words, different mutants may not necessarily alter initiation sequence preference, but instead may shift the initiation probability of use in a polar fashion within some set of already usable start sites. In this view, Pol II initiation efficiency may cooperate with a directional scanning process that has its own rate. Pol II with increased catalytic activity enables initiation earlier within the scanning window, increasing catalytic efficiency (initiation probability) of earlier usable start sites usages, and shifting TSS usage distribution upstream.
Additive effects on ADH1 TSS distributions in Pol II-GTF mutant strains indicate that individual defects of each allele are present in the double mutant strains. How might we understand these defects? Defects in sua7 are consistent with defects in initiation efficiency and likely represent reduced TFIIB functions. TFIIB function in concert with the Pol II active site might represent communication between TFIIB and the active site as has been proposed (Sainsbury et al. 2013) or parallel roles for TFIIB and the Pol II active center during putative TSS scanning. We speculate that sua7 Pol II LOF double mutants that have severe growth defects or are lethal as shown in Figure 2 have exacerbated defects in initiation efficiency from those of single mutants, e.g., those shown for sua7-1 (Cho and Buratowski 1999). Furthermore, mutual suppression of TSS defects in GTF-Pol II double mutants is predicted to result from suppression of initiation defects, which might be tested for GTF-Pol II mutant combinations in GTF-dependent biochemical systems for abortive or productive Pol II initiation such as the system used recently by Fishburn and Hahn (2012). Because Pol II alleles are expected to have additional defects in elongation and termination, GTF alleles do not strongly suppress overall growth defects of Pol II mutants even though TSS defects in some cases examined here are strongly suppressed. Our previous work detecting splicing defects as a consequence of Pol II alleles’ presumed altered elongation functions did not detect similar splicing defects for GTF alleles. Defects detected were milder and of opposite polarity for sua7-3 and tfg2∆261-273 strains relative to Pol II alleles with upstream or downstream shifts in TSS distributions, arguing against predictable defects of these alleles in Pol II elongation (Braberg et al. 2013).
tfg2 TSS phenotypes are similar to those of Pol II GOF alleles and raise the question of how alteration of TFIIF function alters TSS distribution. Previous work had indicated that a TFG1 mutant exhibited an increased ability to stimulate Pol II activity in an abortive initiation assay (Khaperskyy et al. 2008). Fishburn and Hahn (2012) recently reported a negative role for TFIIF in suppressing TSS usage at upstream positions of promoters (nearer to a TATA element). Such a negative role is likely balanced by positive requirements for TFIIF activity in promoting initiation. Conceivably, tfg1 and tfg2 mutants have this negative role—possibly an autoinhibitory function of TFIIF that is alleviated during scanning to downstream positions—specifically or selectively compromised. In light of such a model, upstream TSS shifting tfg alleles would confer increased initiation activity, phenocopying Pol II GOF mutants for altered TSS distribution. In in vitro transcription experiments, TFIIF stimulation of abortive initiation was compromised by the Pol II LOF H1085Y allele of the TL (Cabart et al. 2014). TFIIF stimulation of abortive initiation likely represents one of a number of positive TFIIF roles in initiation. If the observed in vitro stimulation were related to upstream TSS shifts at ADH1 observed in tfg alleles, then compromise of the TL might be expected to be epistatic to tfg phenotypes based on biochemical results. Because tfg2 alleles can still alter TSS distribution in Pol II LOF alleles such as H1085Y, it suggests that the biochemical requirement in vitro of TFIIF for a WT TL may be distinct or bypassed by tfg2 alleles studied here.
The mechanism driving S. cerevisiae start site scanning likely derives from a combination of factors, Pol II activity and transcription bubble opening promoted by TFIIH. Assuming that upstream bubble opening, as was observed at GAL1 and GAL10 (Giardina and Lis 1993), is universal at yeast promoters, a major question is, how does the bubble transit to the distal start sites? One possibility is that a large transcription bubble is extended to the start site region. In this case, PICs would need to accommodate extensive single-stranded DNA. A recent cryo-EM structure and model of the yeast PIC appears consistent with such accommodation (Murakami et al. 2013). In this instance, TFIIH might drive extension of the downstream bubble edge through the activity of its Ssl2/Rad25 subunit (the yeast homolog of human ERCC3/XPB). An alternative model in which a smaller region of melted DNA translocates along with the open PIC toward the start region may also be possible, but almost nothing is known of the organization of nucleic acids in such hypothetical complexes or how translocation of the putative bubble would be controlled during initiation. In translocating Pol II elongation complexes, GTFs are not present and interactions between Pol II, both DNA strands, and nascent RNA organize the upstream edge of the transcription bubble.
Genetic analyses have allowed us to examine the relationships between Pol II activity mutants, known initiation factors, and candidates for possible modifiers of Pol II initiation activity. Our experiments indicate that Pol II genetic interactors need not perturb TSS selection, and that initiation defects are likely only a partial driver of Pol II allele growth phenotypes. Altered Pol II activity through TL defects do not bypass or appear epistatic to the alleles of TFIIB or TFIIF studied here for ADH1 TSS selection, suggesting that they each function separately as part of a concerted process to promote efficient TSS selection.
Supplementary Material
Acknowledgments
We thank Steve Hahn (Fred Hutchinson Cancer Research Center) for providing tfg2∆146-180, tfg2∆233-248, tfg2∆261-273 plasmids and Michael Hampsey (Robert Wood Johnson Medical School) for sua7-1 and sua7-3 plasmids. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM097260 and a Welch Foundation Grant A-1763. We thank members of the Kaplan laboratory for helpful discussions and comments regarding the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Welch Foundation.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.015180/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Amberg, D. C., D. Burke, J. N. Strathern, D. Burke, and Cold Spring Harbor Laboratory, 2005 Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Basehoar A. D., Zanton S. J., Pugh B. F., 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709. [DOI] [PubMed] [Google Scholar]
- Basu R. S., Warner B. A., Molodtsov V., Pupov D., Esyunina D., et al. , 2014. Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 289: 24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braberg H., Jin H., Moehle E. A., Chan Y. A., Wang S., et al. , 2013. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154: 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabart P., Jin H., Li L., Kaplan C. D., 2014. Activation and reactivation of the RNA polymerase II trigger loop for intrinsic RNA cleavage and catalysis. Transcription Jan 1: 5. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. S., Hampsey M., 2004. Functional interaction between TFIIB and the Rpb2 subunit of RNA polymerase II: implications for the mechanism of transcription initiation. Mol. Cell. Biol. 24: 3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. T., Hahn S., 2004. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119: 169–180. [DOI] [PubMed] [Google Scholar]
- Chen H. T., Warfield L., Hahn S., 2007. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 14: 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E. J., Buratowski S., 1999. Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 274: 25807–25813. [DOI] [PubMed] [Google Scholar]
- Choi W. S., Yan M., Nusinow D., Gralla J. D., 2002. In vitro transcription and start site selection in Schizosaccharomyces pombe. J. Mol. Biol. 319: 1005–1013. [DOI] [PubMed] [Google Scholar]
- Corden J. L., 2008. Yeast Pol II start-site selection: the long and the short of it. EMBO Rep. 9: 1084–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Armache K. J., Baumli S., Benkert S., Brueckner F., et al. , 2008. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37: 337–352. [DOI] [PubMed] [Google Scholar]
- Dvir A., 2002. Promoter escape by RNA polymerase II. Biochim. Biophys. Acta. 1577: 208–223. [DOI] [PubMed] [Google Scholar]
- Eichner J., Chen H. T., Warfield L., Hahn S., 2010. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29: 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faitar S. L., Brodie S. A., Ponticelli A. S., 2001. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 21: 4427–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT), A. R. Forrest, H. Kawaji, M. Rehli, J. K. Baille et al., 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishburn J., Hahn S., 2012. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol. Cell. Biol. 32: 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-Picos M. A., Krishnamurthy S., Sun Z. W., Hampsey M., 2005. Evidence that the Tfg1/Tfg2 dimer interface of TFIIF lies near the active center of the RNA polymerase II initiation complex. Nucleic Acids Res. 33: 5045–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazy M. A., Brodie S. A., Ammerman M. L., Ziegler L. M., Ponticelli A. S., 2004. Amino acid substitutions in yeast TFIIF confer upstream shifts in transcription initiation and altered interaction with RNA polymerase II. Mol. Cell. Biol. 24: 10975–10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina C., Lis J. T., 1993. DNA melting on yeast RNA polymerase II promoters. Science 261: 759–762. [DOI] [PubMed] [Google Scholar]
- Gleghorn M. L., Davydova E. K., Basu R., Rothman-Denes L. B., Murakami K. S., 2011. X-ray crystal structures elucidate the nucleotidyl transfer reaction of transcript initiation using two nucleotides. Proc. Natl. Acad. Sci. USA 108: 3566–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S., Krishnamurthy S., Hampsey M., 2012. Mechanism of start site selection by RNA polymerase II: interplay between TFIIB and Ssl2/XPB helicase subunit of TFIIH. J. Biol. Chem. 287: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I. H., Aranda A., Proudfoot N., 2000. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 8415–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V., Li N., Hadzhiev Y., Plessy C., Previti C., et al. , 2014. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11: 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Young E. T., 2011. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189: 705–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M., 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62: 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog G. A., Wada T., Handa H., Winston F., 1998. Evidence that Spt4, Spt5, and Spt6, control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12: 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry N. L., Campbell A. M., Feaver W. J., Poon D., Weil P. A., et al. , 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8: 2868–2878. [DOI] [PubMed] [Google Scholar]
- Henry N. L., Bushnell D. A., Kornberg R. D., 1996. A yeast transcriptional stimulatory protein similar to human PC4. J. Biol. Chem. 271: 21842–21847. [DOI] [PubMed] [Google Scholar]
- Hoskins R. A., Landolin J. M., Brown J. B., Sandler J. E., Takahashi H., et al. , 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 21: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull M. W., McKune K., Woychik N. A., 1995. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 9: 481–490. [DOI] [PubMed] [Google Scholar]
- Kaplan C. D., 2013. Basic mechanisms of RNA polymerase II activity and alteration of gene expression in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1829: 39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Holland M. J., Winston F., 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10–GAL7 locus. J. Biol. Chem. 280: 913–922. [DOI] [PubMed] [Google Scholar]
- Kaplan C. D., Larsson K. M., Kornberg R. D., 2008. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol. Cell 30: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. D., Jin H., Zhang I. L., Belyanin A., 2012. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 8: e1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy D. A., Ammerman M. L., Majovski R. C., Ponticelli A. S., 2008. Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Mol. Cell. Biol. 28: 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva M. L., Nedialkov Y. A., Cremona G. H., Purtov Y. A., Lubkowska L., et al. , 2008. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell 30: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus R., Pollock R., Guarente L., 1996. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 15: 1933–1940. [PMC free article] [PubMed] [Google Scholar]
- Kostrewa D., Zeller M. E., Armache K. J., Seizl M., Leike K., et al. , 2009. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462: 323–330. [DOI] [PubMed] [Google Scholar]
- Kuehner J. N., Brow D. A., 2006. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 281: 14119–14128. [DOI] [PubMed] [Google Scholar]
- Larson M. H., Zhou J., Kaplan C. D., Palangat M., Kornberg R. D., et al. , 2012. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc. Natl. Acad. Sci. USA 109: 6555–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Flanagan P. M., Tschochner H., Kornberg R. D., 1994. RNA polymerase II initiation factor interactions and transcription start site selection. Science 263: 805–807. [DOI] [PubMed] [Google Scholar]
- Luse D. S., 2012. Rethinking the role of TFIIF in transcript initiation by RNA polymerase II. Transcription 3: 156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majovski R. C., Khaperskyy D. A., Ghazy M. A., Ponticelli A. S., 2005. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. J. Biol. Chem. 280: 34917–34923. [DOI] [PubMed] [Google Scholar]
- Malagon F., Kireeva M. L., Shafer B. K., Lubkowska L., Kashlev M., et al. , 2006. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics 172: 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Elmlund H., Kalisman N., Bushnell D. A., Adams C. M., et al. , 2013. Architecture of an RNA polymerase II transcription pre-initiation complex. Science 342: 1238724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto I., Ware D. E., Hampsey M., 1992. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68: 977–988. [DOI] [PubMed] [Google Scholar]
- Pinto I., Wu W. H., Na J. G., Hampsey M., 1994. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem. 269: 30569–30573. [PubMed] [Google Scholar]
- Ranish J. A., Hahn S., 1991. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J. Biol. Chem. 266: 19320–19327. [PubMed] [Google Scholar]
- Sainsbury S., Niesser J., Cramer P., 2013. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493: 437–440. [DOI] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L., 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18: 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Weissman J. S., Krogan N. J., 2006. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40: 344–352. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., Winston F., Styles C. A., Fink G. R., 1984. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc. Natl. Acad. Sci. USA 81: 2431–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. W., Hampsey M., 1995. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92: 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. W., Hampsey M., 1996. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 16: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. W., Tessmer A., Hampsey M., 1996. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 24: 2560–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorovskaya O. V., Engel K. L., French S. L., Cui P., Vandeventer P. J., et al. , 2013. Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II. Cell Reports 4: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. H., Pinto I., Chen B. S., Hampsey M., 1999. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. Y., Carson D. J., Ma J., 2002. The role of TFIIB-RNA polymerase II interaction in start site selection in yeast cells. Nucleic Acids Res. 30: 3078–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Degen D., Ho M. X., Sineva E., Ebright K. Y., et al. , 2014. GE23077 binds to the RNA polymerase ’i’ and ’i+1’ sites and prevents the binding of initiating nucleotides. eLife 3: e02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.