Figure 1.
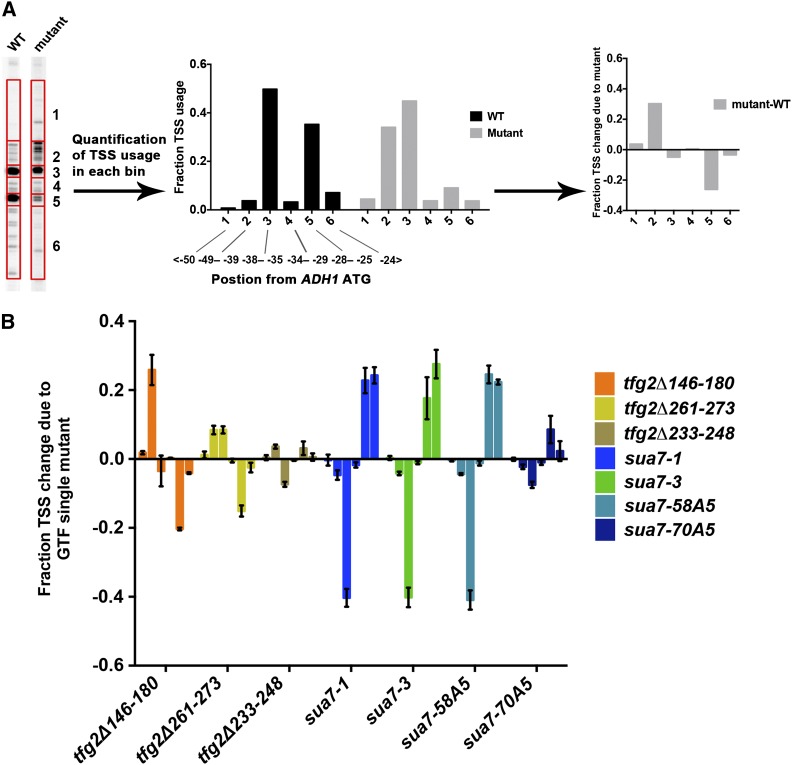
Transcription start site (TSS) usage distribution at ADH1 and its alteration by Pol II GTF mutants. (A) TSSs detected by primer extension at ADH1 are distributed over a range of positions. To quantify TSS distributions, ADH1 start site signals were divided into six bins separated by promoter position and normalized to total signal for each lane (left panel). TSS usage distributions were quantified for different strains (middle panel). Relative change in normalized TSS usage distribution for mutant compared to WT (negative numbers indicate relative decrease in TSS position usage; positive numbers indicate relative increase) is then calculated and plotted (right panel). (B) Alterations in TSS usage at ADH1 caused by each GTF mutant shown were quantified as in (A). Start site defects of these GTF mutants are consistent with previous publications (Wu et al. 1999; Eichner et al. 2010), except for sua7-A5 alleles, which are in contrast to (Zhang et al. 2002). Graphs show average of at least three independent determinations with error bars representing SDs. See Figure S1 for representative primer extension experiments.