Significance
Quality control of proteins is critical to cellular homeostasis. The BCL2-associated athanogene cochaperone 6 (Bag6) complex, which contains Bag6, transmembrane domain recognition complex 35 (TRC35), and ubiquitin-like 4A (Ubl4A), plays an essential role in targeting transmembrane domains either to the endoplasmic reticulum or degradation. Bag6 is a central hub for numerous activities, functionally linked to an array of cellular pathways, from immunoregulation to apoptosis. Here we define the molecular architecture of this heterotrimer, revealing distinct binding sites on Bag6 for TRC35 and Ubl4A. The truncated Bag6 complex defined in this study is sufficient to facilitate substrate transfer from small glutamine-rich tetratricopeptide repeat-containing protein α (SGTA) to TCR40. In addition, structural and biochemical characterization of the BAG domain of Bag6 demonstrate that it is not a canonical BAG domain.
Keywords: GET pathway, Scythe, Bat3, X-ray crystallography, tail-anchored proteins
Abstract
BCL2-associated athanogene cochaperone 6 (Bag6) plays a central role in cellular homeostasis in a diverse array of processes and is part of the heterotrimeric Bag6 complex, which also includes ubiquitin-like 4A (Ubl4A) and transmembrane domain recognition complex 35 (TRC35). This complex recently has been shown to be important in the TRC pathway, the mislocalized protein degradation pathway, and the endoplasmic reticulum-associated degradation pathway. Here we define the architecture of the Bag6 complex, demonstrating that both TRC35 and Ubl4A have distinct C-terminal binding sites on Bag6 defining a minimal Bag6 complex. A crystal structure of the Bag6–Ubl4A dimer demonstrates that Bag6–BAG is not a canonical BAG domain, and this finding is substantiated biochemically. Remarkably, the minimal Bag6 complex defined here facilitates tail-anchored substrate transfer from small glutamine-rich tetratricopeptide repeat-containing protein α to TRC40. These findings provide structural insight into the complex network of proteins coordinated by Bag6.
The well-studied BCL2-associated athanogene cochaperone 6 (Bag6, also known as “BAT3” or “Scythe”) plays a central role in membrane protein quality control, with additional links to apoptosis, gene regulation, and immunoregulation (for reviews, see refs. 1–3). Recent studies demonstrated that Bag6 forms a heterotrimeric Bag6 complex with ubiquitin-like 4A (Ubl4A) and transmembrane domain recognition complex 35 (TRC35) (4, 5) that mediates the fates of membrane proteins in tail-anchor (TA) protein targeting (6), mislocalized protein degradation (7), and endoplasmic reticulum (ER)-associated protein degradation (4). The many roles of the Bag6 complex likely are centered on its ability to bind exposed hydrophobic regions of proteins, such as transmembrane domains. In the cytoplasm, the Bag6 complex directs substrates either to targeting factors for the ER membrane (for TA proteins) or to ubiquitylation and subsequent proteasomal degradation.
Bag6 initially was described as part of the gene cluster that included the human MHC class III on chromosome 6, resulting in its first name, “HLA-B–associated transcript 3” (BAT3) (8). The genomic localization suggested a role in immune response, and this suggestion has been supported by evidence of its roles in Th1 cell survival (9), natural killer cell cytotoxicity (10), and MHC class II molecule presentation (11, 12). The initial Bag6 link to apoptosis was based on its interaction with Reaper, an apoptosis-inducing Drosophila protein (13). Bag6 was not identified in initial searches for functional homologs of Bag1 (14), which identified four additional proteins (Bag2–5) (15). The designation of Bag6 as a Bag family member came later from limited sequence homology to the defined BAG domain and an apparent heat shock cognate 70 (Hsc70)-regulating activity found in other Bag family members (16).
In mammals, Bag6 has been shown to be critical in the targeting of TA proteins to the ER by the transmembrane recognition complex (TRC) pathway (5), a process best understood in the equivalent fungal guided entry of tail-anchored proteins (GET) pathway (17, 18). Although Bag6 is missing in fungi, the analogous yeast complex contains two proteins, Get4 and Get5/Mdy2, which are homologs of the mammalian proteins TRC35 and Ubl4A, respectively. In yeast, these two proteins form a heterotetramer that regulates the handoff of the TA protein from the cochaperone small, glutamine-rich, tetratricopeptide repeat protein 2 (Sgt2) [small glutamine-rich tetratricopeptide repeat-containing protein (SGTA) in mammals] to the delivery factor Get3 (TRC40 in mammals) (19–22). It is expected that the mammalian homologs, along with Bag6, play a similar role (23–27). Bag6 also interacts with other proteins such as apoptosis-inducing factor, glycoprotein 78 (gp78), regulatory particle 5, and brother of regulator of imprinted sites (BORIS) (16, 27–32) and can homo-oligomerize, increasing the level of complexity (30). These findings build a picture of Bag6 as a central hub for a diverse physiological network of proteins.
A variety of diseases, ranging from cancer to autoimmune disorders and diabetes, are linked to Bag6 (33–37). Despite this demonstrated importance, structural characterization of the Bag6 complex is lacking. The longest and most common isoform of the Bag6 gene encodes an 1,132-aa protein (38) with an N-terminal ubiquitin-like (UBL) domain that has been characterized structurally (PDB ID codes 4EEW, 4DWF, and 1WX9), a large proline-rich central domain that is predicted to be unstructured, and a C-terminal predicted BAG domain (Bag6-BAG). In this study, we map the TRC35- and Ubl4A-binding regions to the C terminus of Bag6. Based on these results, the structure of the complex between the heterodimerization domains of Bag6 and Ubl4A was solved, revealing unexpected structural homology to Get5 and showing that the Bag6-BAG is a “mock” BAG domain, as was demonstrated further biochemically. Finally, a defined minimal Bag6 complex facilitates the transfer of a TA substrate from SGTA to TRC40 in vitro.
Results
TRC35 Binds the Bag6 Nuclear Localization Sequence, Whereas Ubl4A Binds the BAG Domain.
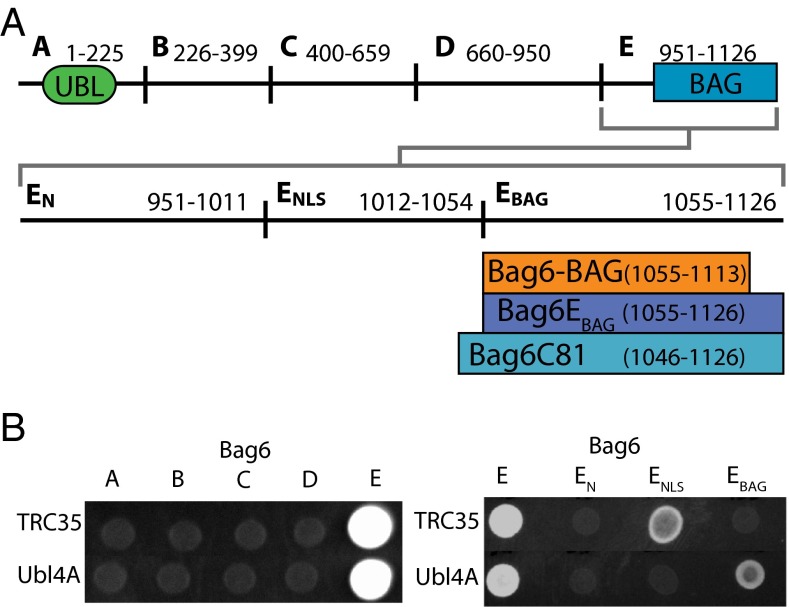
To define the molecular architecture of the heterotrimeric Bag6 complex (Bag6, Ubl4A, and TRC35), a series of yeast two-hybrid assays were performed. Bag6 was divided into five fragments: A (amino acids 1–225), B (amino acids 226–399), C (amino acids 400–659), D (amino acids 660–950), and E (amino acids 951–1,126), with the activating domain attached at the N terminus (Fig. 1A); TRC35 or Ubl4A contained N-terminal DNA-binding domains. TRC35 and Ubl4A both showed a positive interaction with the C-terminal Bag6E fragment that contains the nuclear localization sequence (NLS) and the BAG domain (Fig. 1B). To refine the interfaces, Bag6E was divided further into an N-terminal domain, an NLS domain, and the putative BAG domain (EN, ENLS, and EBAG respectively) (Fig. 1A). TRC35 showed an interaction with ENLS (Fig. 1B) confirming the in vivo result that TRC35 masks the Bag6-NLS, preventing nuclear targeting (4). Ubl4A showed an interaction with EBAG (Fig. 1B), a surprising result because none of the five previously characterized BAG domains was known to form stable interactions with other proteins.
Fig. 1.
Bag6 has distinct binding sites for TRC35 and Ubl4A at its C terminus. (A) Scheme of the five different Bag6 fragments and of the subfragments of Bag6E, which is divided further into the N terminus (EN), the NLS (ENLS), and a fragment containing only the putative BAG domain (EBAG). (B, Left) Yeast two-hybrid assay between Bag6 fragments and either TRC35 or Ubl4A. The A fragment contains the UBL domain, and the E fragment contains the NLS and putative BAG domain. (Right) Yeast two-hybrid assay of TRC35 or Ubl4A and the Bag6 E fragments.
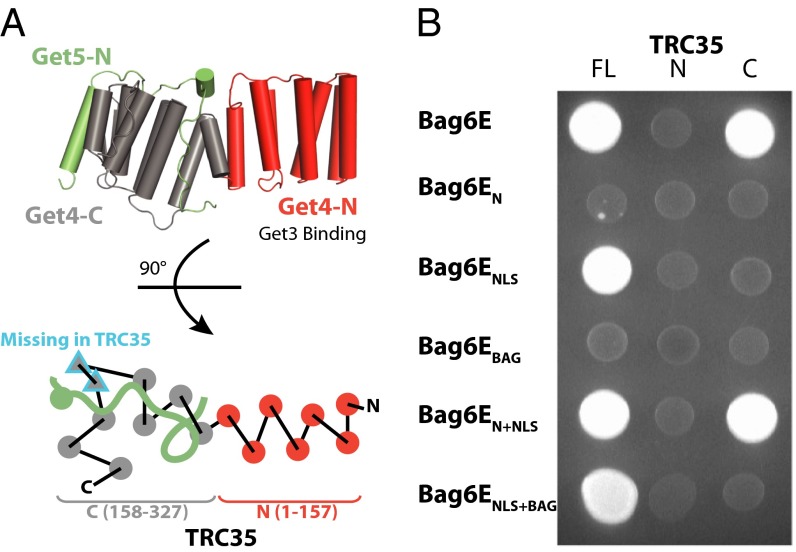
According to sequence alignment, Ubl4A lacks the Get5 N-terminal domain, and TRC35 lacks the β-loop in Get4 (Fig. 2A), both of which are involved in the Get4 interface with Get5 (19). This difference suggests there are different interactions in the Bag6 complex. One possibility is that the region around the Bag6 NLS acts structurally like the Get5 N domain by binding the C domain of TRC35. To confirm this hypothesis, a two-hybrid experiment was performed with TRC35 split into either an N domain (TRC35-N, residues 1–157) or a C domain (TRC35-C, residues 158–327), as had been done previously for Get4 (Fig. 2A) (39). As predicted, Bag6E showed a clear interaction with TRC35-C and no interaction with TRC35-N (Fig. 2B). Surprisingly, the smaller Bag6ENLS did not interact with TRC35-C (Fig. 2B) despite the previously seen interaction with full-length TRC35 (Fig. 1B). This interaction was restored with the longer Bag6EN,NLS, suggesting that additional contacts are required to form a stable interaction. This extended region defines a minimal complex with TRC35 bound to the C terminus of Bag6 in close proximity to Ubl4A, similar to the architecture found in the yeast Get4-5 complex.
Fig. 2.
Defining the interaction between Bag6 and TRC35 by yeast two-hybrid assay. (A, Upper) Diagram of the Get4/Get5-N complex (PDB ID code 3LKU). Get4 is shown in gray and red, and Get5 is shown in green. (Lower) A top-view schematic representation of the architecture. The two β-strands in Get4 that are missing in TRC35 are outlined in blue. Ubl4A does not contain a Get5-N equivalent. (B) Bag6E fragments containing the activating domain were combined with full-length TRC35 (FL), TRC35-N (residues 1–157), or TRC35-C (residues 158–327) conjugated to the binding domain. TRC35-N and TRC35-C were defined by sequence alignment with Get4.
The next goal was to purify the heterotrimeric complex. Both full-length Bag6 and TRC35 were recalcitrant to recombinant expression in Escherichia coli. For TRC35, expression required removal of residues at the N and C termini that are not conserved, TRC35(23–305). This truncated TRC35 behaved like wild-type by yeast two-hybrid assay with Bag6E (Fig. S1A). Additionally, based on all the interaction results, a minimal Bag6 fragment (Bag6min, residues 1,001–1,126) was constructed that removed the 50 N-terminal residues of Bag6E. The coexpression of Bag6min, TRC35(23–305), and Ubl4A resulted in a stable complex that could be purified (Fig. S1B). Although a TRC35–Bag6E complex could not be obtained, the heterodimeric Ubl4A–Bag6E was well expressed and could be purified to homogeneity.
The Crystal Structure of Bag6-BAG/Ubl4A-C.
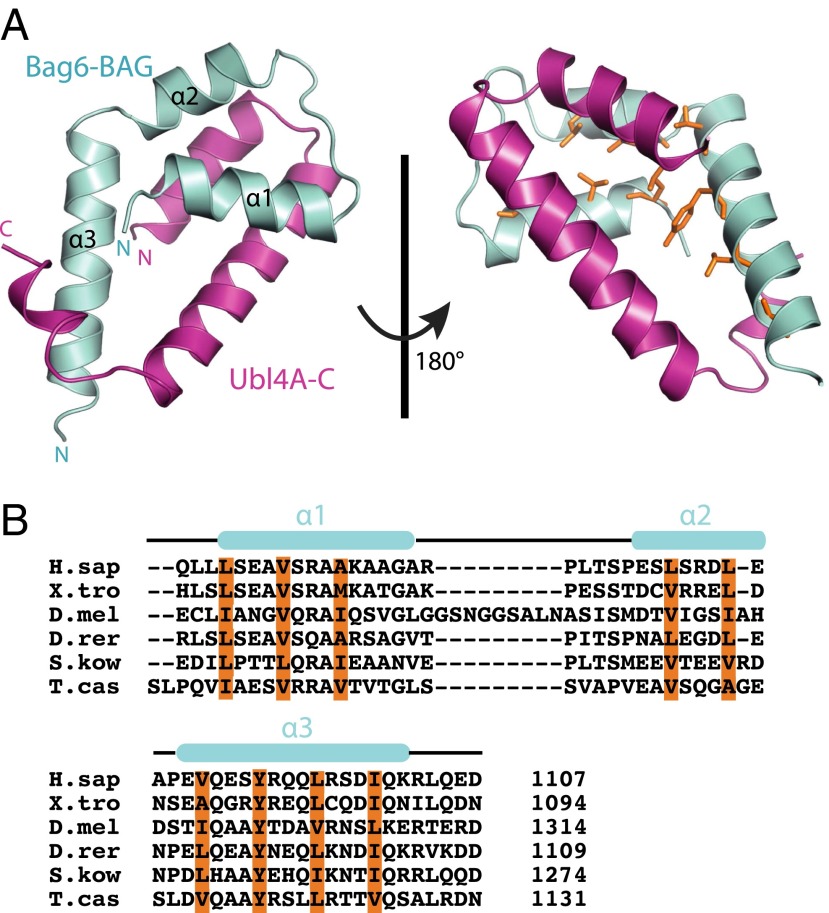
For structural characterization, multiple variants of the Bag6min complex were pursued for crystallization. One consisting of the Ubl4A C-terminal dimerization domain (Ubl4A-C) and the Bag6 BAG domain (Bag6-BAG, residues 1,054–1,107) resulted in well-formed crystals. A complete 2.1-Å native dataset was collected in the space group P21 and was phased using an iodide derivative. The final refinement resulted in an Rfree of 28.0% (Table S1). Both domains are primarily helical (Fig. 3A) with an extensive dimer interface dominated by conserved hydrophobic residues that results in 2,485 Å2 of buried surface (Fig. 3A). Bag6-BAG contains three helices in an extended conformation making few intramolecular contacts. Ubl4A-C contains three helices with the first two forming an interface and the short third helix wrapping around Bag6-BAG (Fig. 3A). All the conserved hydrophobic residues of Bag6-BAG participate in dimerization (Fig. 3B).
Fig. 3.
The crystal structure of the Bag6-BAG/Ubl4A-C heterodimer. (A) The overall structure of the Bag6-BAG/Ubl4A-C heterodimer in ribbon representation with Bag6-BAG shown in cyan and Ubl4A-C shown in magenta. Hydrophobic residues in Bag6 involved in packing are highlighted as orange sticks. (B) Sequence alignment of Bag6-BAG. D.mel, Drosophila melanogaster; D.rer, Danio rerio; H.sap, Homo sapiens; S.kow, Saccoglossus kowalevskii; T.cas, Tribolium castaneum; and X.tro, Xenopus tropicalis. The secondary structure based on the structure is highlighted above the text. The conserved hydrophobic and aromatic residues involved in the hydrophobic packing interactions between Bag6-BAG and Ubl4A-C are highlighted in orange. The extended Drosophila melanogaster sequence is a predicted protein sequence based on theoretical translation and may not reflect a physiological isoform.
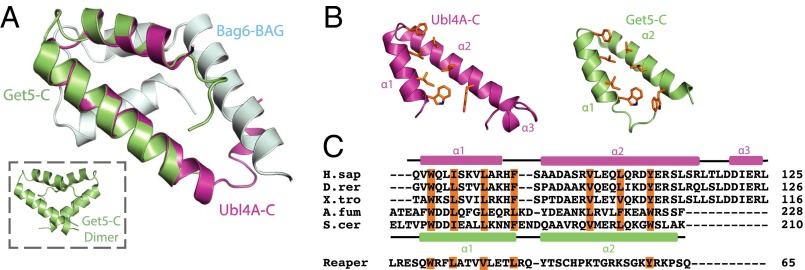
Although fungal Get5 forms a stable homodimer mediated by Get5-C, Ubl4A alone is primarily a monomer (20). Because Ubl4A forms a heterodimer with Bag6, one might expect a novel fold for Ubl4A-C; instead, the Ubl4A-C heterodimerization domain has a structure identical to the Get5-C homodimerization domain, an rmsd of 0.94 Å for equivalent backbone residues (Fig. 4 A and B). The hydrophobic residues that form the core in Get5 homodimerization—W179, I182, L186, F190, V200, L204, W208—are conserved in Ubl4A—W96, I99, L103, F107, V115, L119, Y123 (Fig. 4 B and C).
Fig. 4.
Ubl4A-C and Get5-C are structurally homologous. (A) A monomer of Get5-C (green) (PDB ID code 3VEJ) and Ubl4A-C (pink) are overlaid. Bag6-BAG is included in cyan. (Inset) the Get5-C homodimer. (B) Ubl4A-C and Get5-C are juxtaposed with conserved residues involved in dimerization highlighted as orange sticks. (C) Sequence alignment of Ubl4A-C homologs (A.fum, Aspergillus fumigatus; D.rer, Danio rerio; H.sap, Homo sapiens; S.cer, Saccharomyces cerevisiae; X.tro, Xenopus tropicalis) and the Drosophila apoptosis-inducing protein Reaper. The Ubl4A-C and Get5C secondary structures are shown above (pink) and below (green), respectively. Conserved hydrophobic residues involved in dimerization are highlighted in orange.
Bag6-BAG Is a Mock BAG Domain.
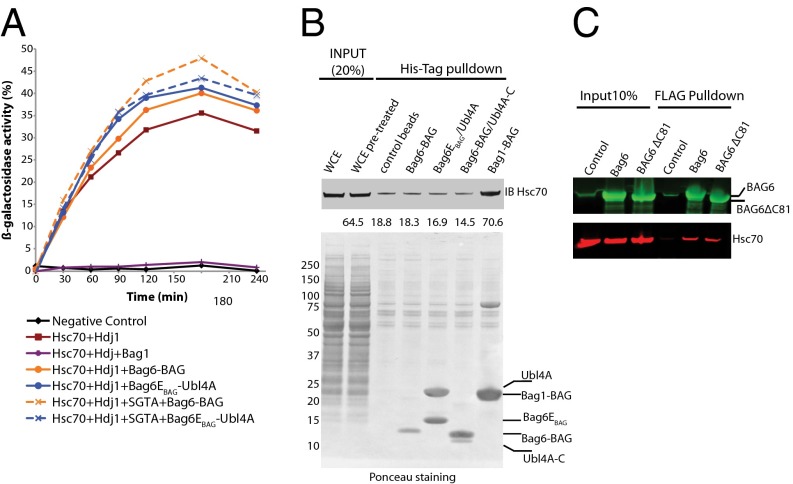
Previous results had demonstrated that the ability of Bag6 to inhibit Hsc70 refolding of substrates in vitro is dependent on the presence of the 81 C-terminal residues (Bag6-C81), which include the BAG domain (16). This result was suggested to be equivalent to results for the Bag1 BAG domain, a demonstrated nucleotide-exchange factor for Hsc70 (40). If true, Bag6-BAG should inhibit Hsc70-mediated protein folding (41). To assay this possibility, denatured β-galactosidase was folded in vitro in the presence of human Hsc70 and human DnaJ protein I (Hdj1), as done previously (16, 42). Folding was measured as the percent of β-galactosidase activity recovered after the folding reaction was quenched. With both Hsc70 and Hdj1 present, a maximal refolded activity of ∼35% was recovered after 180 min (Fig. 5A, brown line), but no refolding was seen when only BSA was added (Fig. 5A, black line). The addition of human Bag1-BAG to Hsc70 and Hdj1 completely inhibited the ability of Hsc70 to fold the protein (Fig. 5A, purple line), as was consistent with previous results (41). Conversely, the addition of Bag6-BAG had no effect on refolding by Hsc70 (Fig. 5A, solid orange line).
Fig. 5.
Bag6-BAG is not a canonical BAG domain. (A) Hsc70-mediated refolding of β-galactosidase in the presence of Bag1-BAG (purple trace), Bag6-BAG (solid orange trace), Bag6EBAG/Ubl4A (solid blue trace), SGTA and Bag6-BAG (dashed orange trace), SGTA and Bag6EBAG/Ubl4A (dashed blue trace), or BSA (black trace) as a negative control. (B) Affinity-tagged Bag6-BAG, Bag6EBAG/Ubl4A, Bag6-BAG/Ubl4A-C, or Bag1-BAG was loaded onto cobalt resin beads and incubated with 293T whole-cell extract (WCE). Eluted samples were immunoblotted with Hsc70 antibody (Upper) and were Ponceau stained (Lower). (C) FLAG-Bag6 or FLAG-Bag6ΔC81 was overexpressed in 293T cells, and anti-FLAG resin was used to capture them with bound factors. Bag6 antibody on the blot is detected in green, and Hsp70 antibody is detected in red.
The inconsistency with previous results might be explained by copurification of endogenous Bag6-binding partners. In the earlier study, affinity-tagged full-length human Bag6 and Bag6ΔC81 were expressed and purified from insect cells over a single-affinity resin (16). Because the proteins are highly conserved, it is reasonable to assume that endogenous insect Ubl4A could be a contaminant and might have contributed to the inhibition. The introduction of the Bag6-BAG/Ubl4A complex to the reaction had no effect on folding (Fig. 5A, blue line). SGTA, a cochaperone, recently has been shown to form a specific complex with Ubl4A (24) and also may have been present in the endogenously purified sample. SGTA had no significant effect on activity (Fig. S2) with or without the other factors (Fig. 5A, dashed lines). Bag6-C81, which was required for inhibition in the previous study, is slightly larger than the BAG domain defined here (Fig. 1A). However, using this larger fragment also had no effect on folding (Fig. S2). Together, these results suggest that, although the 81 C-terminal residues of Bag6 are required for its chaperoning activity, Bag6-C81 does not act as a bona fide BAG domain to cooperate with Hsc70.
This conclusion was supported further by binding assays. A 6x-histidine (6xHis)-tagged Hsc70-nucleotide binding domain (NBD), the expected binding site for Bag proteins, was incubated with purified Bag6-C81, Bag6-BAG, Bag6-BAG/Ubl4A, or Bag1-BAG. After incubation, Hsc70-NBD was captured on nickel-nitrilotriacetic acid-agarose (Ni-NTA) beads along with any associated protein. As expected, Hsc70-NBD was able to capture the Bag1-BAG domain (Fig. S3C). On the other hand, Hsc70-NBD was unable to capture any of the Bag6 C-terminal fragments at levels above background. Thus, one would conclude that Bag6-BAG is unlike canonical BAG domains in its ability to interact with Hsc70 in vitro.
Although the evidence makes it clear that the Bag6 BAG domain does not interact with Hsc70 in isolation, the role of other unknown factors could not be ruled out. For instance, SGTA could mediate the interaction between Bag6-BAG and Hsp70, because it binds Hsp70 via its tetratricopeptide repeat domain and Ubl4A via its N terminus (25). To address the possibility of accessory factors, tagged variants of BAG domains were incubated with 293T whole-cell lysate (Fig. 5B). The positive control Bag1-BAG again was able to capture a significant amount of Hsc70. A complex with Bag6-BAG and either the crystallized fragment or full-length Ubl4A was unable to capture Hsc70.
These results are in contrast to previous experimental results in which Bag6 inhibition of Hsc70 was dependent on the presence of the BAG domain (16), but could not rule out the possibility of binding Hsc70 in the context of the full-length Bag6. To test this possibility, 293T cells were transfected with either FLAG-Bag6 or FLAG-Bag6ΔC81 and were captured with anti-FLAG resin. Proteins bound to the beads were blotted with both Bag6 and Hsp70 antibodies (green and red, respectively, in Fig. 5C). Full-length Bag6 captured a small amount of Hsc70 over background; however, the lack of the previously annotated BAG domain (Bag6ΔC81) had no effect on the amount of captured Hsc70 (Fig. 5C).
Based on the structural characterization, the Bag6-BAG domain is shorter (47 residues) than canonical BAG domains (76–112 residues), and the three helices do not form a BAG-like three-helix bundle; the few residues equivalent to those involved in Hsp70 binding of other BAG domains have different orientations (Fig. S3A). Furthermore, circular dichroism of Bag6-BAG alone indicates no stable secondary structure (Fig. S4). These results imply that a primary role for Bag6-BAG is to heterodimerize with Ubl4A, because the two are found in a stoichiometric complex (5, 7). Combined, these results suggest that Bag6-BAG, both structurally and biochemically, is not a true BAG domain but instead is a mock BAG domain.
The Bag6min Complex Is an Independent Module That Facilitates TA Handoff.
The fungal Get4-5 complex binds ATP-bound Get3 and inhibits its ATPase activity, priming Get3 for TA substrate capture from Sgt2 (22, 43). One would expect that the trimeric Bag6 complex, which contains the mammalian Get4-5 orthologs, regulates TRC40 in a similar manner. To test this notion, an in vitro assay was developed to probe the role of the Bag6min complex in TA handoff from SGTA to TRC40 using recombinantly purified proteins (the scheme is shown in Fig. 6A). When coexpressed, fungal Sgt2 binds GET-dependent TA substrates, and this complex can be purified (21). Here, histidine-tagged SGTA (hSGTA) was coexpressed in E. coli with the yeast TA protein Sec61 beta homolog (Sbh1) that contained an N-terminal maltose-binding protein (MBP–Sbh1), resulting, after a two-step purification, in a stable hSGTA/MBP–Sbh1 complex (Fig. S1B). The final component, TRC40, was generated as a N-terminal GST tag (GST-TRC40) (Fig. S1B), as done previously (44, 45). Transfer was initiated by incubation of hSGTA/MBP–Sbh1 either with GST-TRC40 alone or with the Bag6min complex (Fig. 6A). The samples then were precipitated with anti-GST resin, washed in two steps in the absence of ATP, and probed after Western blotting with both MBP and GST antibodies. GST-TRC40 was able to capture some TA from SGTA alone, as seen previously for the yeast system (Fig. S5 A and C) (21). The additional presence of the Bag6min complex resulted in an ATP-dependent increase in TA transfer to TRC40 (Fig. S5A, compare lanes 4 and 6). This increase in transfer was not a result of a bridged capture of TRC40 pulling down TA still bound to SGTA. The interaction between SGTA and Ubl4a is predicted to have very fast off-rates (25), and SGTA would be removed rapidly during the wash steps. Moreover, capture of TA by TRC40 was insensitive to increasing salt concentration (Fig. S5 B and D), despite the SGTA/Ubl4a and TRC35/TRC40 interactions being dominated by electrostatics, the latter also requiring ATP (22, 25). The Bag6min fragment used in the study does not contain the Bag6 substrate-binding domain (23); therefore, the Bag6 complex can promote the handoff of substrate from SGTA to TRC40 without Bag6 engaging the substrate directly.
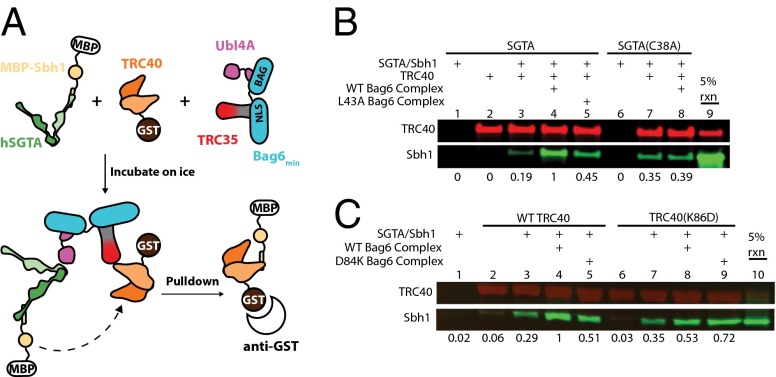
Fig. 6.
The Bag6min complex facilitates TA transfer from SGTA to TRC40. (A) The in vitro TA handoff reaction scheme. Recombinantly purified hSGTA-MBP/Sbh1 complex was incubated with GST-TRC40 and the indicated recombinant proteins. After incubation on ice for 10 min, GST-TRC40 and bound substrate were precipitated with anti-GST resin followed by three washing steps and Western blotting. (B) Mutants affecting SGTA binding to the Bag6min complex reduce TA transfer to TRC40. GST-TRC40 was captured on anti-GST resin after incubation in the presence of ATP with SGTA/MBP-Sbh1 or SGTA(C38A)/MBP-Sbh1 alone or with the Bag6min or Bag6min/Ubl4A(L43A) complex. Eluted samples were immunoblotted with anti-GST (red) and anti-MBP antibody (green) and then were quantified by Odyssey Infrared Imaging System analysis software. Relative values of captured Sbh1 are shown below each lane with the experiment containing all wild-type components as the reference. Sbh1 fluorescence values were normalized for each trial based on GST-TRC40 captured in each lane. Values are averages of six independent experiments. SDs are included in Fig. S5. The 5% rxn lane corresponds, in all cases, to loading 5 µL of the wild-type reaction before capture. (C) Regulatory mutant GST-TRC40(K86D) and TRC35(D84K)Bag6min complexes were incubated with the indicated recombinant proteins and ATP. GST-TRC40(K86D) and bound factors were captured and analyzed as in B.
SGTA binds the Bag6 complex via the UBL domain of Ubl4A; consequently, the Bag6-dependent handoff should require this interaction (24, 25). To test this hypothesis, the mutants hSGTA(C38A) and Ubl4A(L43A) were generated; previously, the equivalent mutations in yeast were shown to disrupt the homologous interaction (25). As expected, each mutation resulted in a similar loss of substrate handoff relative to wild type (Fig. 6B, compare lanes 4, 5, and 8, and Fig. S5E). This result highlights the importance of this interaction for the bridging by Bag6 during TA transfer.
In yeast, Get4-5 binding regulates Get3 ATPase activity and TA targeting (43). Therefore, in addition to the bridging role of the Bag6 complex, it is critical to test if this larger complex plays a regulatory role in TA targeting. The recently published structure of yeast Get4 bound to Get3 highlighted a regulatory interface separate from the binding interface (22). When residues on either side of this regulatory interface were mutated (the charge swaps K69D on Get3 and D74K on Get4), each mutation resulted in a loss of ATPase inhibition, reduction of TA insertion into microsomes, and a loss of fitness in vivo, despite maintaining a stable complex in vitro (22). Combining the charge swap mutants restored the Get4 regulatory activity (22). For TRC40, the corresponding regulatory mutation, K86D, resulted in a reduction of the Bag6 complex-facilitated handoff (Fig. 6C, compare lanes 4 and 8, and Fig. S5F). Similarly, the corresponding regulatory mutation in TRC35, D84K, resulted in a reduction of facilitated handoff (Fig. 6C, compare lanes 4 and 5, and Fig. S5F). Excitingly, as seen for the yeast system (22), the combination of these two charge swap mutants resulted in a rescue of the facilitated handoff (Fig. 6C, lane 9, and Fig. S5F). These results show that this minimal Bag6 complex acts as an independent TA-targeting module and performs a regulatory role similar to that of the fungal Get4-5 complex despite the different architectures.
Discussion
The Bag6 complex has been implicated in various cellular pathways, necessitating a description of its molecular architecture. In this study, we examined the trimeric complex and determined the high-resolution structure of the complex between the heterodimerization domains of Bag6 and Ubl4A. Our structural and functional characterization revealed that Bag6-BAG is a mock BAG domain and does not act independently as a BAG protein-like nucleotide-exchange factor with Hsc70. Instead, it acts as part of a scaffold, bridging TRC35 and Ubl4A, resulting in a minimal Bag6 complex that regulates TA handoff.
The demonstrated inability of Bag6-BAG to influence Hsc70 activity is unsurprising if one considers sequence comparisons with canonical BAG domains. This result brings into question the reported inhibition of Hsp70-mediated β-galactosidase refolding by Bag6 (16). A possible explanation is the holdase activity of Bag6. Bag6 prevents the aggregation of unfolded luciferase by forming a stable interaction with the exposed hydrophobic core, preventing Hsp70-mediated refolding (4). Similar Bag6 holdase activity may have prevented refolding of β-galactosidase. Deletion of the 81 C-terminal residues may disrupt this holdase through an unknown mechanism, possibly by occluding the binding site on the truncated Bag6. The observed Hsp70 inhibition by Bag6, therefore, would be a result of sequestration of the unfolded substrate by Bag6 via its hydrophobic substrate-binding region, which also could be recognized by Hsp70, resulting in the interaction between Bag6 and Hsp70.
The first functional annotation of Scythe (the Bag6 Xenopus homolog) was an ability to bind Reaper, an apoptosis-inducing protein in Drosophila, thus inhibiting Reaper-induced apoptosis in Xenopus oocyte extracts (13, 46). Reaper induces apoptosis in a variety of model systems, including Xenopus oocyte extract (47), SF-21 insect cells (48), and HeLa human cancer cells (49). Sequence alignment of the conserved Ubl4A dimerization domain and Reaper reveals the conservation of most of the residues involved in Bag6-BAG/Ubl4A-C dimerization (Fig. 4C), suggesting that Reaper may disrupt the Bag6/Ubl4A interaction. This hypothesis would be consistent with Reaper’s binding the 312-residue C-terminal truncation of Scythe (ScytheC312), leading to the release of bound factors (13).
Although full-length Scythe and Bag6 inhibit Reaper- and ricin-triggered apoptosis, excess ScytheC312 (13) or the 131 C-terminal residues (50) of Bag6 can induce apoptotic events. Because the C-terminal fragment would be consistent with the Bag6min complex defined here, excess Bag6 C-terminal residues would disconnect the triaging and holdase/degradation roles of the complex. The apoptosis connection then could be linked to TA targeting. Overexpression of Bag6 in HeLa cells exposed to ricin, an apoptosis inducer, leads to an increase in endogenous Bcl-2 protein levels, whereas Bag6 knockdown causes down-regulation of Bcl-2 proteins (50). Several proteins that belong to the Bcl-2 family, including Bcl-2, MCL1, BAX, and BOK, are tail-anchored and reside both at the ER and the mitochondria (51–53). The Bag6 complex then would play an important role in regulating the localization and turnover of these Bcl-2 proteins, and this role could be disrupted by Bag6 cleavage.
These results support a model in which the primary role of the Bag6 C terminus is to bridge TRC35 and Ubl4A. Possible Bag6 dimerization would form a heterohexamer, creating a complex analogous to the Get4-5 heterotetramer found in yeast and providing strong mechanistic parallels in TA targeting (Fig. S6). The N-terminal UBL domain of Bag6 connects the proteasome, where it interacts with RP non-ATPase 10c (29), with the ER, where it interacts with gp78 and ubiquitin regulatory X domain-containing protein 8 (4, 30). Downstream of this connection, the proline-rich domain has been implicated as the holdase domain binding to exposed hydrophobic regions and polyubiquitinated defective ribosomal products (4, 23, 54). Bag6 then acts as a scaffolding protein, simultaneously binding ubiquitylation machinery, the proteasome, TA-targeting factors, and proteins to be triaged. The molecular details of its decision-making process and how this process relates to its other functions in apoptosis, gene regulation, and immunoregulation are important questions for future studies. The results presented in this paper provide an important foundation for understanding this intriguing complex.
Materials and Methods
Detailed descriptions of experiments are provided in SI Materials and Methods. Briefly, PJ69-4α strain yeast was cotransformed with pGAD and pGBDU, and protein–protein interaction was assessed by its ability to grow in SC-Ura-Leu-Ade plates. For crystallization, folding assays, and Hsc70-NBD pulldowns, human genes were subcloned, expressed in E. coli, and purified using Ni-affinity, anion exchange, and size-exclusion chromatography. A truncated Bag6-BAG fragment and Ubl4A-C were crystallized, and the structure was determined using single wavelength anomalous dispersion. All the capture assays from whole-cell extract were performed with 293T cell extracts. FLAG-tagged recombinant full-length Bag6 and Bag6ΔC81 were recombinantly purified from 293T cells. All proteins used in the TA handoff assay were purified in E. coli.
Supplementary Material
Acknowledgments
We thank Daniel Lin and Jens Kaiser for help with data processing; Yoko Shibata and Richard Morimoto (Northwestern University) for plasmids; Michael Rome and Meera Rao for critical reading of the manuscript; members of the W.M.C. laboratory for support and useful discussions; the staff at the Advanced Light Source for assistance with synchrotron data collection; and Gordon and Betty Moore for support of the Molecular Observatory at California Institute of Technology. W.M.C. is supported by National Institutes of Health Grant R01GM097572.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank, www.rcsb.org/pdb/ (accession no. 4WWR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402745112/-/DCSupplemental.
References
- 1.Lee JG, Ye Y. Bag6/Bat3/Scythe: A novel chaperone activity with diverse regulatory functions in protein biogenesis and degradation. BioEssays. 2013;35(4):377–385. doi: 10.1002/bies.201200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawahara H, Minami R, Yokota N. BAG6/BAT3: Emerging roles in quality control for nascent polypeptides. J Biochem. 2013;153(2):147–160. doi: 10.1093/jb/mvs149. [DOI] [PubMed] [Google Scholar]
- 3.Binici J, Koch J. BAG-6, a jack of all trades in health and disease. Cell Mol Life Sci. 2014;71(10):1829–1837. doi: 10.1007/s00018-013-1522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42(6):758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466(7310):1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Hessa T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475(7356):394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spies T, Bresnahan M, Strominger JL. Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B. Proc Natl Acad Sci USA. 1989;86(22):8955–8958. doi: 10.1073/pnas.86.22.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangachari M, et al. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3–mediated cell death and exhaustion. Nat Med. 2012;18(9):1394–1400. doi: 10.1038/nm.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pogge von Strandmann E, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity. 2007;27(6):965–974. doi: 10.1016/j.immuni.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Kämper N, et al. γ-Interferon-regulated chaperone governs human lymphocyte antigen class II expression. FASEB J. 2012;26(1):104–116. doi: 10.1096/fj.11-189670. [DOI] [PubMed] [Google Scholar]
- 12.Pai RK, Askew D, Boom WH, Harding CV. Regulation of class II MHC expression in APCs: Roles of types I, III, and IV class II transactivator. J Immunol. 2002;169(3):1326–1333. doi: 10.4049/jimmunol.169.3.1326. [DOI] [PubMed] [Google Scholar]
- 13.Thress K, Henzel W, Shillinglaw W, Kornbluth S. Scythe: A novel reaper-binding apoptotic regulator. EMBO J. 1998;17(21):6135–6143. doi: 10.1093/emboj/17.21.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takayama S, et al. Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell. 1995;80(2):279–284. doi: 10.1016/0092-8674(95)90410-7. [DOI] [PubMed] [Google Scholar]
- 15.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274(2):781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 16.Thress K, Song J, Morimoto RI, Kornbluth S. Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 2001;20(5):1033–1041. doi: 10.1093/emboj/20.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chartron JW, Clemons WM, Jr, Suloway CJ. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22(2):217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12(12):787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chartron JW, Suloway CJ, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA. 2010;107(27):12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chartron JW, VanderVelde DG, Rao M, Clemons WM., Jr Get5 carboxyl-terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J Biol Chem. 2012;287(11):8310–8317. doi: 10.1074/jbc.M111.333252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40(1):159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gristick HB, et al. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol. 2014;21(5):437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leznicki P, et al. The association of BAG6 with SGTA and tail-anchored proteins. PLoS ONE. 2013;8(3):e59590. doi: 10.1371/journal.pone.0059590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Cai M, Yang Y, Huang L, Ye Y. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Reports. 2012;2(6):1633–1644. doi: 10.1016/j.celrep.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chartron JW, VanderVelde DG, Clemons WM., Jr Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Reports. 2012;2(6):1620–1632. doi: 10.1016/j.celrep.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leznicki P, High S. SGTA antagonizes BAG6-mediated protein triage. Proc Natl Acad Sci USA. 2012;109(47):19214–19219. doi: 10.1073/pnas.1209997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long P, Samnakay P, Jenner P, Rose S. A yeast two-hybrid screen reveals that osteopontin associates with MAP1A and MAP1B in addition to other proteins linked to microtubule stability, apoptosis and protein degradation in the human brain. Eur J Neurosci. 2012;36(6):2733–2742. doi: 10.1111/j.1460-9568.2012.08189.x. [DOI] [PubMed] [Google Scholar]
- 28.Desmots F, Russell HR, Michel D, McKinnon PJ. Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J Biol Chem. 2008;283(6):3264–3271. doi: 10.1074/jbc.M706419200. [DOI] [PubMed] [Google Scholar]
- 29.Kikukawa Y, et al. Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J. 2005;272(24):6373–6386. doi: 10.1111/j.1742-4658.2005.05032.x. [DOI] [PubMed] [Google Scholar]
- 30.Xu Y, Liu Y, Lee JG, Ye Y. A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation. J Biol Chem. 2013;288(25):18068–18076. doi: 10.1074/jbc.M112.449199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen P, et al. BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol Cell Biol. 2008;28(21):6720–6729. doi: 10.1128/MCB.00568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akahane T, Sahara K, Yashiroda H, Tanaka K, Murata S. Involvement of Bag6 and the TRC pathway in proteasome assembly. Nat Commun. 2013;4:2234. doi: 10.1038/ncomms3234. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh YY, et al. Human lymphocyte antigen B-associated transcript 2, 3, and 5 polymorphisms and haplotypes are associated with susceptibility of Kawasaki disease and coronary artery aneurysm. J Clin Lab Anal. 2010;24(4):262–268. doi: 10.1002/jcla.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harney SM, et al. Fine mapping of the MHC Class III region demonstrates association of AIF1 and rheumatoid arthritis. Rheumatology (Oxford) 2008;47(12):1761–1767. doi: 10.1093/rheumatology/ken376. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degli-Esposti MA, et al. Ancestral haplotypes reveal the role of the central MHC in the immunogenetics of IDDM. Immunogenetics. 1992;36(6):345–356. doi: 10.1007/BF00218041. [DOI] [PubMed] [Google Scholar]
- 37.Vandiedonck C, et al. Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci USA. 2004;101(43):15464–15469. doi: 10.1073/pnas.0406756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerji J, Sands J, Strominger JL, Spies T. A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc Natl Acad Sci USA. 1990;87(6):2374–2378. doi: 10.1073/pnas.87.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang YW, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285(13):9962–9970. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sondermann H, et al. Structure of a Bag/Hsc70 complex: Convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291(5508):1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- 41.Takayama S, et al. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16(16):4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15(12):2969–2979. [PMC free article] [PubMed] [Google Scholar]
- 43.Rome ME, Rao M, Clemons WM, Shan SO. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc Natl Acad Sci USA. 2013;110(19):7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124(Pt 8):1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto Y, Sakisaka T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell. 2012;48(3):387–397. doi: 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 46.Thress K, Evans EK, Kornbluth S. Reaper-induced dissociation of a Scythe-sequestered cytochrome c-releasing activity. EMBO J. 1999;18(20):5486–5493. doi: 10.1093/emboj/18.20.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans EK, et al. Reaper-induced apoptosis in a vertebrate system. EMBO J. 1997;16(24):7372–7381. doi: 10.1093/emboj/16.24.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vucic D, Seshagiri S, Miller LK. Characterization of reaper- and FADD-induced apoptosis in a lepidopteran cell line. Mol Cell Biol. 1997;17(2):667–676. doi: 10.1128/mcb.17.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tait SW, Werner AB, de Vries E, Borst J. Mechanism of action of Drosophila Reaper in mammalian cells: Reaper globally inhibits protein synthesis and induces apoptosis independent of mitochondrial permeability. Cell Death Differ. 2004;11(8):800–811. doi: 10.1038/sj.cdd.4401410. [DOI] [PubMed] [Google Scholar]
- 50.Wu YH, Shih SF, Lin JY. Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J Biol Chem. 2004;279(18):19264–19275. doi: 10.1074/jbc.M307049200. [DOI] [PubMed] [Google Scholar]
- 51.Strasser A. The role of BH3-only proteins in the immune system. Nat Rev Immunol. 2005;5(3):189–200. doi: 10.1038/nri1568. [DOI] [PubMed] [Google Scholar]
- 52.Szegezdi E, Macdonald DC, Ní Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am J Physiol Cell Physiol. 2009;296(5):C941–C953. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- 53.Echeverry N, et al. Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ. 2013;20(6):785–799. doi: 10.1038/cdd.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minami R, et al. BAG-6 is essential for selective elimination of defective proteasomal substrates. J Cell Biol. 2010;190(4):637–650. doi: 10.1083/jcb.200908092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.