Fig. 3.
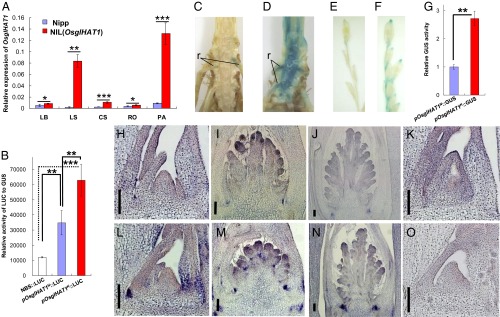
Altered OsglHAT1 promoter activity underlies the QTL effect on grain weight regulation. (A) qPCR analysis of OsglHAT1 expression pattern. RNA was isolated and quantitated by qPCR, normalized to ubiquitin. CS, culm tissue containing shoot apical meristem; LB, leaf blade; LS, leaf sheath; PA, young panicle; RO, root. (B) Transient assay using maize leaf protoplasts to test OsglHAT1 promoter activity. GUS staining of transgenic samples containing pOsglHAT1N::GUS (C and E) and pOsglHAT1K::GUS construct (D and F). r, root hair. (G) Quantification of the GUS signal that harbors the construct as indicated. In situ RNA hybridization of OsglHAT1 shows expression in the vegetative stage (H and L) and during the reproductive stage (I, J, M, and N); (K and O) Negative controls of OsglHAT1 in situ RNA hybridization that uses a sense probe. (Scale bars: 100 μm.) The length of the promoters pOsglHAT1N and pOsglHAT1K used in B–G was 1,681 and 1,652 bp, respectively, upstream of the ORF of OsglHAT1 alleles. Sample sections in H–K are Nipp genotypes, and in L–O are Kasa genotypes. *P < 0.1; **P < 0.05; ***P < 0.001. Student’s t test was used to generate the P values.
