Figure 3.
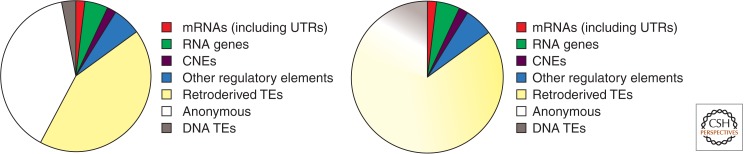
The functional versus nonfunctional human genome. The pies represent the various segments of the genome applying more conservative estimates. mRNAs (∼1.3% accounting for ORFs) including their 5′- and 3′-UTRs (∼0.8%) occupy slightly >2% of the human genome. All other RNAs (non-protein-coding RNAs) are estimated to cover about 5% (green), maximally 10%. This figure is under debate. Likewise, there is a conservative estimate for regulatory elements of ∼8%. Other sources argue up to 20% (Bernstein et al. 2012; Shen et al. 2012). Because conserved non-protein-coding DNA elements (CNEs, ∼2%, purple) often encode cis-regulatory elements (Hiller et al. 2012), the other regulatory elements are shown as 6% only (blue). TEs derived from RNA intermediates contribute ∼43% (yellow) and DNA transposons only 3% (brown). The remainder (39%) corresponds to anonymous, scrambled sequences (white). The estimated 26% introns (Bernstein et al. 2012) occupy mostly the yellow and white segments; intronic regulatory elements and encoded RNAs (e.g., snoRNAs, miRNAs) are accounted for in the respective segments. The pie on the right is identical, except that the retroposons (yellow) and, to a lesser extent, the DNA transposons (brown), blend into the anonymous sequence (white) to reflect TE ancestry of the nondescript sequences as well.
