Figure 3.
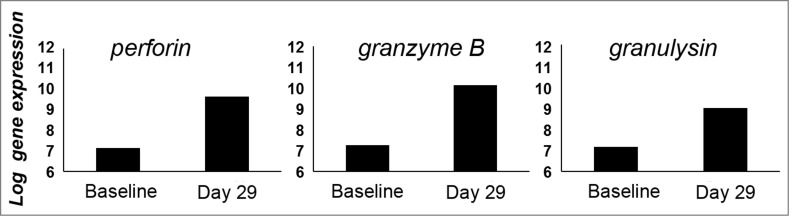
Total RNA was extracted from snap-frozen core needle tumor biopsies taken at baseline and 1 mo after the treatment initiation and gene expression profiling was performed by using HumanHT-12 Illumina microbead chips (Illumina Inc., San Diego, CA USA). The average signal values were quantile-normalized, log2-transformed, and annotated using the package lumi (Bioconductor open source software). Chip-dependent batch-effects were removed using empirical Bayes methods. Markedly elevated expression levels of genes encoding cytotoxic factors perforin, granulysin and granzyme B in post-treatment sample suggest that CD8+ TILs show an effector phenotype.
