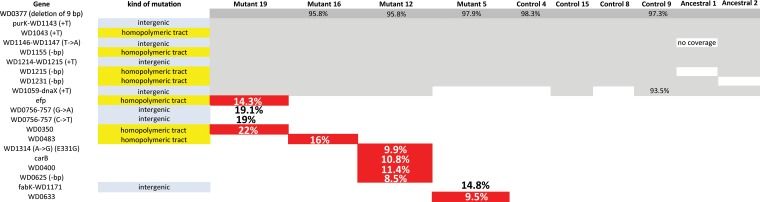
FIG 3.
Mutations in passaged Wolbachia within chic221/+ background or control flies. Whole-genome sequencing using an Illumina MiSeq sequencer was used to identify individual mutations in each of 4 lines from chic221/+ passaged Wolbachia and compared to 4 control passaged Wolbachia as well as the ancestral stock (sequenced twice). The identified loci affected by mutation are listed, and the presence of a mutation within each lineage is depicted. If the change is predicted to alter the amino acid encoded at that position, the mutation is colored in red. If the mutation was found in <100% of the mapped reads, the percentage is shown.