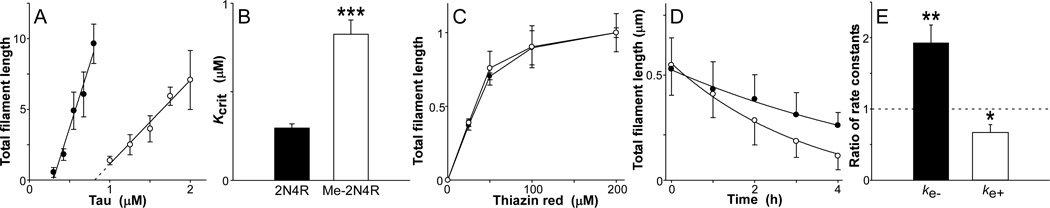
Figure 5. Lys methylation modulates tau filament extension rates.
(A) Unmodified (solid circle) or 5.5 mol/mol methylated tau (hollow circle) were incubated (18 h at 37°C) at varying bulk concentrations in the presence of 100 µM Thiazine red inducer, then assayed for filament formation by electron microscopy (reported in arbitrary units). Total filament lengths were then plotted against bulk protein concentration, where each data point represents the mean ± SD of triplicate determinations and the solid lines represent best fit of the data points to linear regression. The abscissa intercept was obtained by extrapolation (dotted lines) and taken as the critical concentration (Kcrit). (B) Replot of data from panel (A), where each bar represents the Kcrit ± propagated SEE. Methylation increased Kcrit nearly 3-fold relative to unmodified tau (***, p < 0.001). (C) Unmodified (solid circle) or 5.5 mol/mol methylated tau (hollow circle) were incubated (18 h at 37°C) at constant supersaturation (i.e., 0.5 µM above Kcrit) in the presence of varying concentrations of Thiazine red, and then assayed for filament formation by electron microscopy. Each data point represents total filament lengths/field from triplicate determinations (reported in arbitrary units). (D) Filaments prepared from unmodified (solid circle) and 5.5 mol/mol methylated tau (hollow circle) as described above were diluted below Kcrit in assembly buffer, and the resultant disaggregation followed as a function of time by electron microscopy. Each data point represents total filament length per field ± SD (n = 3), whereas the solid line represents best fit of data points with Eq. 2. The first-order decay constant kapp was estimated from each regression and used in conjunction with filament length (shown in figure) and number at time t = 0 to calculate dissociate rate constant ke−. ke+ was then obtained from Eq. 3. (E) Replot of data from panel (D), where each bar represents the ratio of rate constants for filament extension (ke+) and dissociation (ke−) determined for 5.5 mol/mol methylated versus unmodified tau ± propagated SEE. A ratio of one, corresponding to no difference in rate, is marked by the dashed line. Methylation increased filament dissociation while decreasing filament extension. *, p < 0.05; **, p < 0.01 for comparison of methylated versus unmodified rate constants.