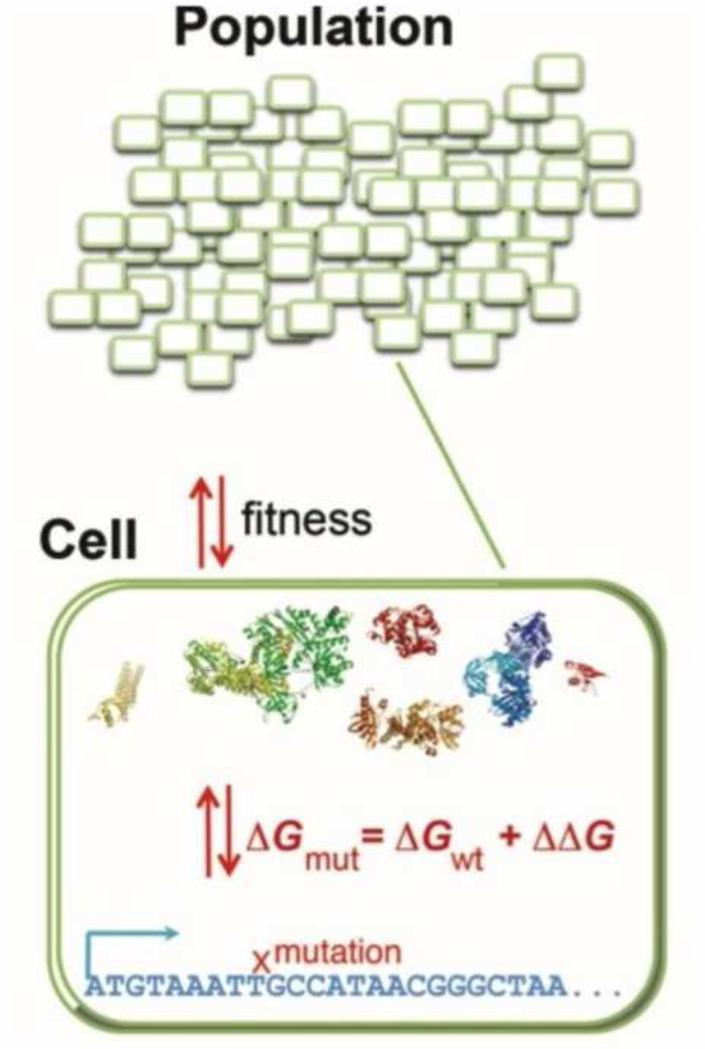
Figure 1. Schema of a bottom-up and multi-scale evolutionary models.
A model population consists of N organisms each with explicit genomes that encode proteins. The fitness of an organism is proportional to the folding stability of the proteins in the cytoplasm. The protein products are represented by their 3D structures from the protein databank (PDB)[16*]. When a random mutation occurs in the genome, tools in protein engineering and the 3D structure are used to estimate its effect on folding stability and, consequently, fitness. Alternatively, the proteins can be represented by 3D lattice models that allows for the exact calculation of biophysical properties [65] or for the possibility of a change in fold. The entire population is subject to mutation, drift, and purifying selection.