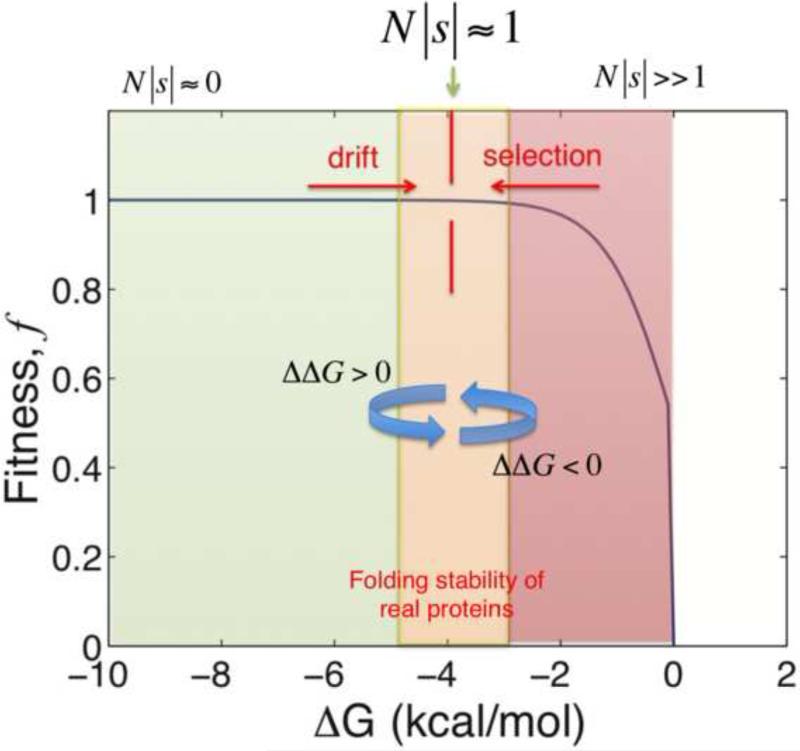
Figure 2. Fitness effects of mutations on a protein folding thermodynamic landscape.
The integrated biophysics-population dynamics models typically assume that the fitness of the model organism is proportional to the total number of folded (functional) proteins in the cytoplasm. That is, fitness f ∝1/(1+eβΔG). Under this assumption, equation 2 defines how molecular changes (ΔΔG) map to fitness effect (s) [11,12,15]. In the regime of very stable proteins, the factor eβΔG → 0, thus N|s| ≈ 0 even if ΔΔG values are nonzero. Additionally, because arising mutations are predominantly destabilizing, most mutations that fix in this regime are destabilizing giving rise to a mutational drift of ΔG towards the less stable regime. Conversely, in the regime of unstable proteins, N|s| > > 1 and selection dominates. Hence, in the unstable regime, mutations that fix are predominantly stabilizing. Mutation-selection balance occurs at the folding stability value where N|s| ≈ 1. Altogether, the epistatic interactions mutations on the thermodynamic fitness landscape results in the near neutrality of the fitness effects of fixed substitutions even if their molecular effects (ΔΔG) are non-neutral.