Figure 2.
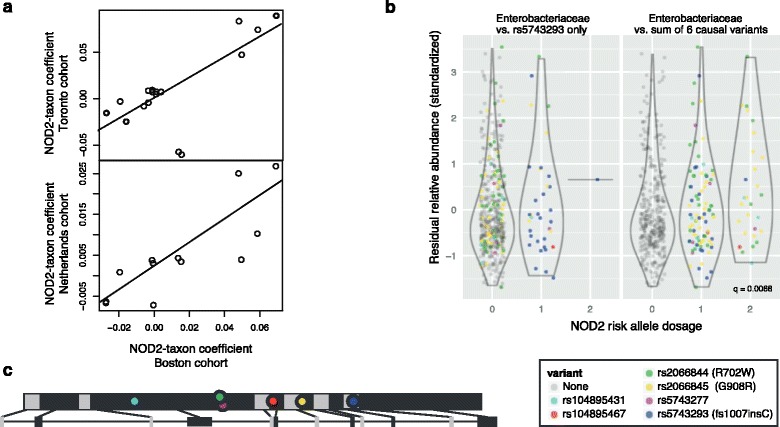
NOD2 fine mapping reveals association with taxonomic and metabolic dysbiosis. (a) Scatterplot of NOD2-bacterial taxon regression coefficients in one study versus the corresponding regression coefficients in another study. We included only those taxa with a nominally significant (P < 0.05) association in a least one of the cohorts being compared. (b) Comparison of residual distributions of Enterobacteriaceae with and without incorporating the six independent known causal NOD2 variants; considering variant rs5743293, only 6.3% of subjects have one or more risk alleles; aggregating risk allele counts across the six variants increases this to 21.8%, and reveals much stronger associations with the microbiome. The strip charts and violin plots show the distribution of standardized residual relative abundance of Enterobacteriaceae versus NOD2 risk allele dosage after data transformation and regression on clinical covariates. Violin plots show the conditional density of residual relative abundance within each dosage level. (c) Relative positions of six NOD2 variants in NOD2 exons [29].
