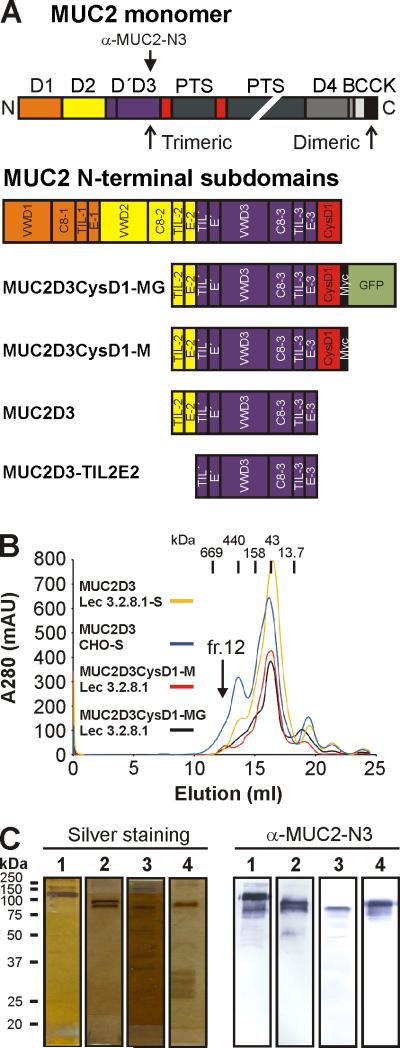
Fig. 1.
Domain structure and purification of MUC2D3 proteins. (A) MUC2 monomer consists of multiple domains: D (D1 (orange), D2 (yellow), D'D3 (blue) and D4 (dark grey)), CysD (red), PTS (dark green), VWB (grey), VWC (light grey) and CK domain (black). The D domains of MUC2 are composed of a defined set of subdomains: VWD1, C8-1, TIL-1 and E-1 (for D1); VWD2, C8-2, TIL-2 and E-2 (for D2); TIL’ and E’ (for D’); VWD3, C8-3, TIL-3 and E-3 (for D3). The three MUC2D3 protein constructs contained remnants of the D2 assembly, namely, TIL-2 (half the subdomain), E-2 and the remaining D’ and D3 assemblies of the D'D3 domains. The MUC2D3CyD1-MG construct included CysD1, Myc tag (black) and GFP (light green) whereas MUC2D3 did not include them and MUC2D3CysD1-M was lacking GFP. The anti-MUC2-N3 antibody recognizes the D'D3 domain. (B) The different MUC2D3 constructs were transfected into CHO-Lec 3.2.8.1, CHO-Lec 3.8.2.1-S and/or CHO-S cells and culture media collected for subsequent protein purification by anion exchange and gel filtration chromatography. All MUC2D3 protein variants were eluting from the gel filtration column in fraction 12 and had an estimated size of ~600 kDa. The elution positions of the gel filtration standards are indicated by vertical bars: thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), ovalbumin (43 kDa) and ribonuclease A (13.7 kDa), respectively. (C) Analysis of MUC2D3 protein variants from fraction 12 after gel filtration by reducing SDS/PAGE, silver staining and western blotting using an affinity-purified rabbit anti-MUC2-N3 antibody: MUC2D3CysD1-MG CHO-Lec 3.2.8.1 (lane 1), 130 kDa; MUC2D3CysD1-M CHO-Lec 3.2.8.1 (lane 2), 100 kDa; MUC2D3 CHO-Lec 3.2.8.1-S (lane 3), 90 kDa; MUC2D3 CHO-S (lane 4), 100 kDa. The migration positions of the molecular weight standards are indicated.