Figure 2. M4 helix is necessary for AMPAR tetramerization.
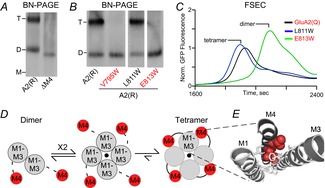
A, blue-native PAGE (BN-PAGE) illustrating that the deletion of M4 (ΔM4) yields only dimers in contrast to wild-type GluA2(R) (A2(R)). Tetramer (T) and dimer (D) bands are based on apoferritin and NativeMark standards. B, example BN-PAGE for single Trp (W) substitutions of the M4 helix in GluA2(R). Trp substitutions at certain positions (V795W and E813W) had the same effect on tetramerization as the deletion of the entire M4, while others (L811W) had no effect. C, fluorescent size-exclusion chromatography (FSEC) of GluA2(Q)–enhanced green fluorescent protein (EGFP) with single M4 substitutions correlated well with results from BN-PAGE. Peaks corresponding to different oligomeric species are indicated. Data from Salussolia et al. (2013). D, putative model for iGluR assembly at the level of the transmembrane domain where a pair of dimers forms a tetrameric complex, presumably stabilized by interactions between the M4 of a subunit and M1–M3 of an adjacent subunit. E, positions in the M4 segment where single Trp substitutions block tetramerization are located along a specific face of the helix (red), referred to as ‘VLGAVE’ (V, valine; L, leucine; G, glycine; A, alanine; V, valine; E, glutamate). The VLGAVE face of the M4 segment from one subunit (dark grey) fits into a groove formed by M1 and M3 from an adjacent subunit (light grey).
