Figure 2.
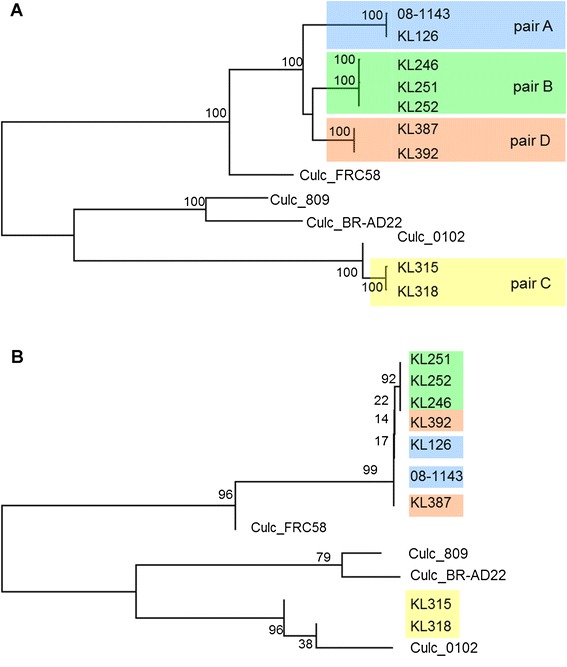
Resequencing reveals zoonotic transmission of C. ulcerans and improves resolution in phylogeny compared with multi-locus sequence typing. (A) Whole genome sequence phylogenetic analysis of the C. ulcerans isolates. The evolutionary history was inferred using the neighborhood joining method [55]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches [56]. The isolates within the pairs are indistinguishable from each other in the dendrogram, indicating very close relationship or even identity, while the isolates of other pairs are clearly separated (B) Phylogenetic analysis for seven MLST loci as in [57]. The phylogenetic analysis was conducted as in Figure 2A. KL251, KL252, KL392, KL126, 08-1143 and KL 387 fall together into one cluster which offers no information on the substructure (bootstrap values 14 to 19), showing that the resolution of MLST is not high enough to sort the isolates into the three pairs as in Figure 2A.
