Figure 2.
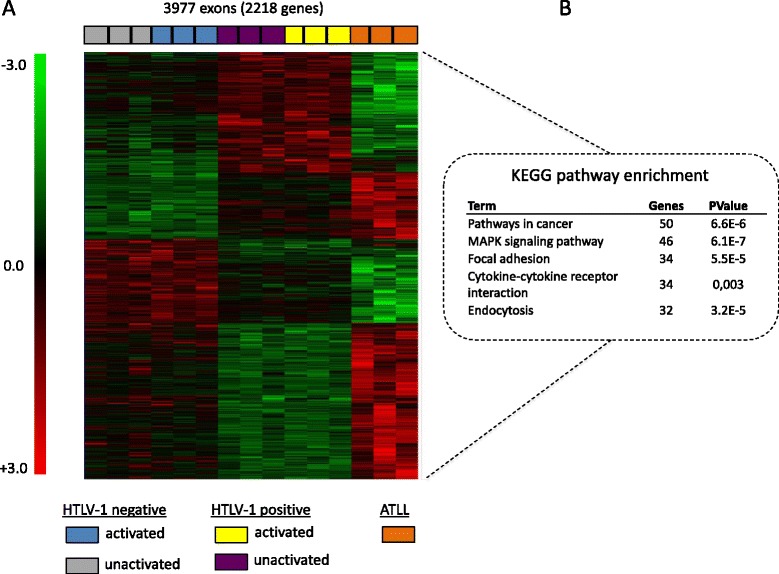
Exon-based hierarchical clustering. Hierarchical clustering analysis was performed with Mev4.0 software (http://www.tm4.org/) using the gene-normalized exon intensities. (A) The selection of exons useful for differentiating between the 15 cell samples was statistically analyzed using a Kruskal-Wallis ANOVA with p < 0.05 representing statistical significance. (B) GO analysis. Top pathways are presented. The gene set corresponded to 2000 genes that displayed the highest splicing index (SI) values. The complete set of genes featured in microarrays (42304 Ensembl geneIDs) was used as a reference background. Official gene symbol annotations are presented in Additional file 5: Table S3.
