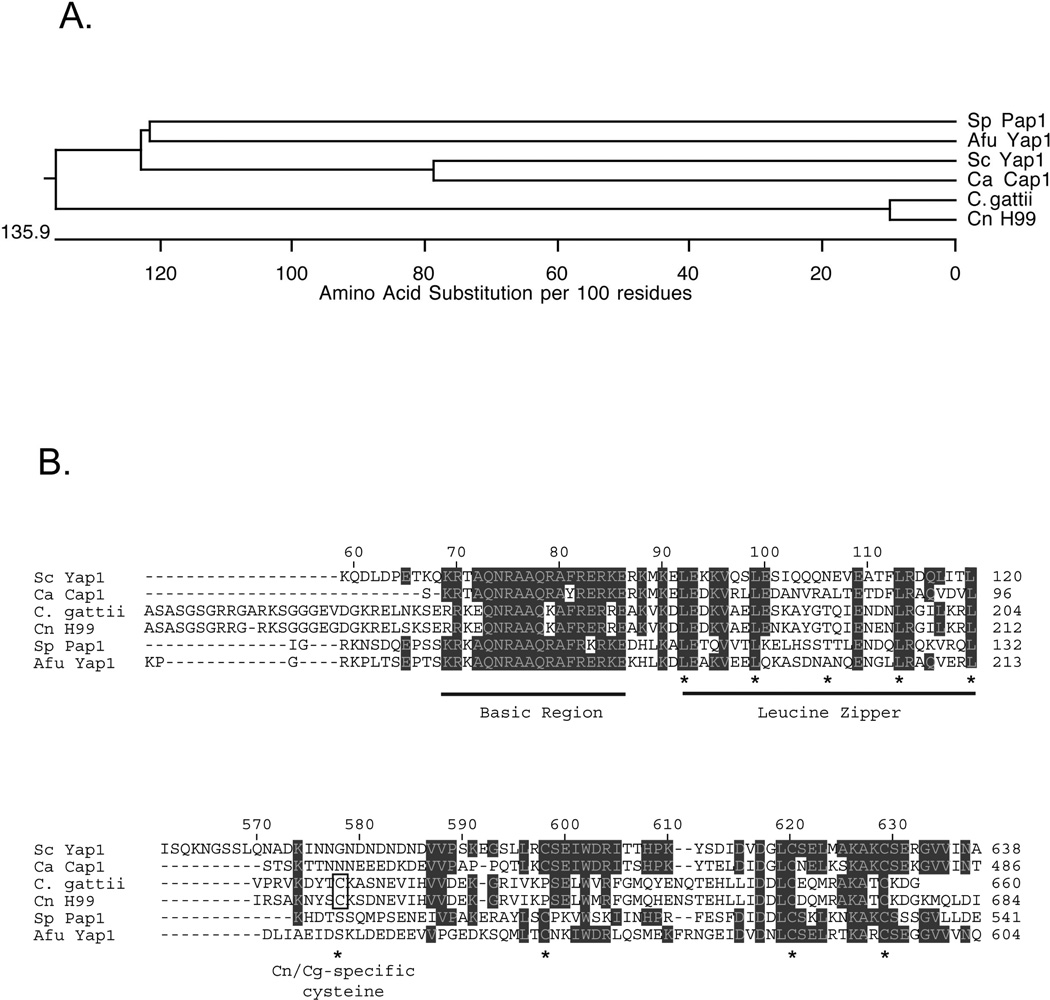
Figure 1. Cryptococcal species encode a homologue of Sc Yap1.
A. An evolutionary tree diagram is shown illustrating the relatedness of different Yap1 homologues from both pathogens and nonpathogens. Sp: Schizosaccharomyces pombe; Afu: Aspergillus fumigatus; Sc: Saccharomyces cerevisiae; Ca: Candida albicans; C. gattii: Cryptococcus gattii; Cn: Cryptococcus neoformans. Note that the C. albicans Yap1 homologue is referred to as Cap1. B. Amino acid sequence alignments from the proteins described above. The numbers at the top refer to the amino acid position along the Sc Yap1 protein chain while the numbers at the right indicate position in each factor. Residues that are in shaded boxes are conserved in at least 3 different proteins. The top panel represents the basic region-leucine zipper domain of each factor with each section of this DNA-binding domain indicated and underlined. Asterisks denote the positions of the canonical leucine residues in the leucine zipper (see (64) for a review). The bottom panel represents the C-terminal cysteine-rich domain that is involved in control of nuclear localization (in all species where it has been examined). The asterisks indicate positions where cysteine residues are found. Note that the most amino-terminal cysteine is in a different position in the Cryptococcus species compared to the others. This unique cysteine residue is indicated by the open box and labeled Cn/Cg-specific cysteine.