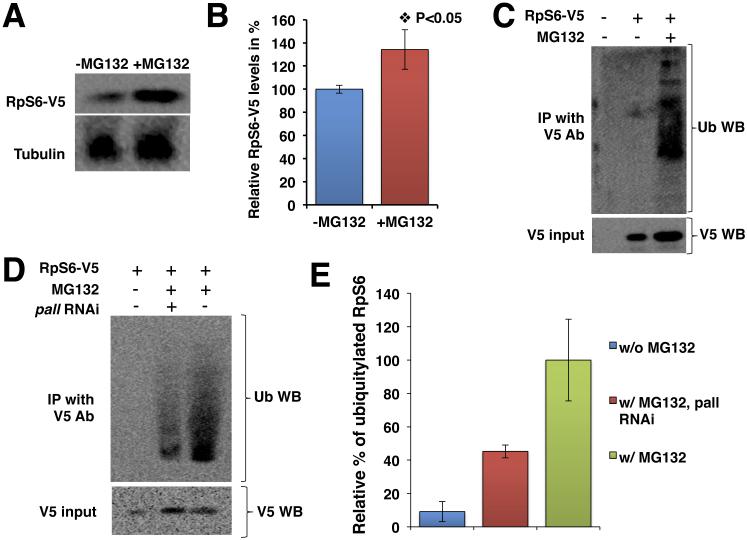
Figure 2. RpS6 is a substrate of PALL for poly-ubiquitylation and proteasomal degradation.
(A) HA-PALLFL stable S2 cells transfected with RpS6-V5 and treated with or without MG132, followed by WB with V5 Ab. The Tubulin WB serves as a loading control. (B) Graph summarizing the quantification of (A). Bars represent the relative percentage when compared to untreated (-MG132) cells ± standard errors from the mean (SEM) of three independent experiments. (C) HA-PALLFL stable S2 cells transfected with RpS6-V5 and Act5C-Ub constructs and treated with or without MG132. V5 Ab immunoprecipitates of RpS6 blotted with Ub Ab to detect poly-ubiquitylated forms of RpS6. (D) Same experiments as in (C) but with or without pre-treating the S2 cells with pall RNAi prior to MG132 treatment and IP with V5 Ab and WB with Ub Ab. In C-D, the input of RpS6-V5 protein is shown by WB of crude cell extracts with V5 Ab. (E) Graph showing the relative quantification of the mean % ± SEM of ubiquitylated RpS6 in S2 cells without MG132 (w/o MG132) and pall-RNAi cells in the presence of MG132 (w/MG132 pall RNAi) when compared to control S2 cells with MG132 (w/MG132).