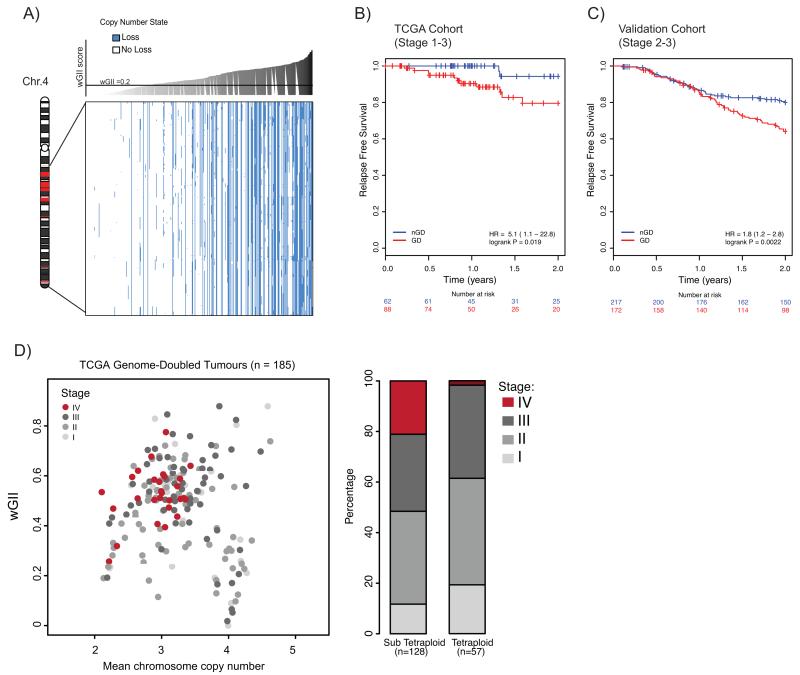
Figure 4.
A) Relationship between copy number loss and wGII in TCGA cohort for genes identified as recurrently lost in tetraploid clones (see Supplementary Table 1). Blue represents loss and white represents no loss (relative to ploidy). CRC tumours (columns) are ordered according to increasing wGII score, from left to right. Every gene (rows) shows a significant correlation between copy number loss and wGII, even when taking into account an increased likelihood of loss in high wGII tumours (P<0.001; see Methods). Chromosome schematic shows where genes reside on chromosome 4; genes within regions shown in red are not depicted in the plot as they are not recurrently lost in all tetraploid clones.
B) Kaplan Meier relapse free survival curves, censored at 2 years for genome-doubled (GD; red) and non-genome doubled (nGD; blue) TCGA cohort CRC tumours (n=150). P=0.019, log-rank test (for full survival curves see Supplementary Fig. 8A).
C) Kaplan Meier relapse-free survival curves, censored at 2 years for genome-doubled (GD; red) and non-genome doubled (nGD; blue) validation cohort CRC tumours (n=389). P=0.0022, log-rank test (for full survival curves see Supplementary Fig.8B).
D) Relationship between wGII, ploidy and tumour stage in genome-doubled tumours. Each circle represents one genome-doubled tumour. The barplot shows the proportion of different tumour stages for tetraploid and sub-tetraploid samples. P=0.0062, Cochrane-Armitage test for trend.