FIG 2.
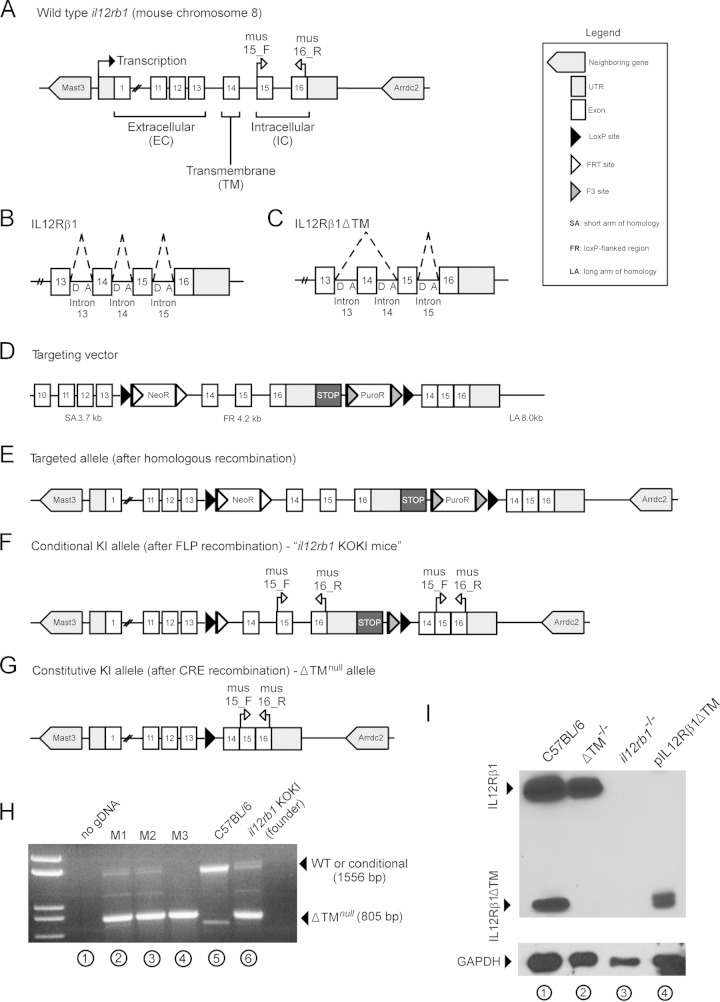
IL12Rβ1ΔTM-deficient mice retain the ability to produce IL12Rβ1 but are unable to produce IL12Rβ1ΔTM. (A) Wild-type il12rb1 is located on mouse chromosome 8, comprises a 5′ UTR, 16 exons, and a 3′ UTR, and is neighbored by the genes mast3 and arrdc2. The first 13 exons of il12rb1 encode the extracellular domains of IL12Rβ1, while exon 14 encodes the transmembrane (TM) domain, and exons 15 and 16 encode intracellular signaling domains. (B and C) The pre-mRNA produced by il12rb1 transcription is normally spliced to produce two distinct isoforms, IL12Rβ1 and IL12Rβ1ΔTM. (B) IL12Rβ1 is produced by inclusion of all il12rb1 exons, including exons 13 to 16; this requires joining of the splice donor “D” and acceptor “A” sites depicted by the dashed lines. (C) IL12Rβ1ΔTM is produced by inclusion of every il12rb1 exon but exon 14. This requires joining of the intron 13 splice D and intron 14 A sites, as depicted by dashed lines. Upon joining the intron 13 “D” and intron 14 “A” sites, exon 14 is removed from the il12rb1 pre-mRNA. This effectively results in “skipping” of exon 14. To generate mice that are unable to produce IL12Rβ1ΔTM but able to produce IL12Rβ1, the wild-type il12rb1 allele (A) was replaced with a targeted allele that, following Cre-recombination, is nonpermissive to il12rb1 pre-mRNA exon 14 skipping (E). Specifically, recombination between il12rb1 and the short arm (SA) and long arm (LA) of a targeting vector was used to introduce a loxP-flanked region (FR) containing the targeted allele, as well as two resistance cassettes (NeoR and PuroR) (D). (F) After FLP recombination, a conditional knockin (KI) allele was generated that, similar to the wild-type allele, was permissive to pre-mRNA exon 14 skipping (i.e., contained the necessary donor and acceptor splice sites). Transcriptional readthrough past the floxed region was prevented via a transcriptional STOP cassette. The mice containing this conditional KI allele are referred to as “il12rb1 KOKI mice.” This conditional allele was removed (G) after crossing mice onto the EIIA-Cre background, leaving a constitutive KI (i.e., ΔTMnull) allele that lacked the introns necessary for exon 14 skipping, rendering mice IL12Rβ1ΔTM deficient. (H) PCR genotyping with the primers mus15_F and mus16_R was used to discriminate mice harboring either a wild-type il12rb1 allele or conditional KI allele and those homozygous for the ΔTMnull allele (i.e., ΔTM−/− mice). Shown is a representative gel image demonstrating the results of genotyping il12rb1 KOKI mice (lane 6), C57BL/6 mice (lane 5), three F2 progeny (lanes 2 to 4), and a “no gDNA control” (lane 1). Indicated in panels A, F, and G are the relative positions of primers mus15_F and mus16_R along the wild-type (A), conditional KI (F), and ΔTMnull (G) alleles. On the sides of the gel in panel H are indicated the expected amplicons from either the wild-type or conditional KI allele amplification (1,556 bp), as well as the ΔTMnull allele amplification (805 bp). (I) To confirm that ΔTM−/− mice are unable to produce IL12Rβ1ΔTM, total cell lysates from C57BL/6 (lane 1), ΔTM−/− (lane 2), and il12rb1−/− ConA blasts were probed via Western blot with anti-IL12Rβ1. Cell lysates from NIH 3T3 cells transfected with pIL12Rβ1ΔTM served as a positive control for IL12Rβ1ΔTM expression (lane 4). All protein lysates were probed with anti-GAPDH as a loading control (bottom panel).