Figure 4. Identification and validation of miR-9 targets.
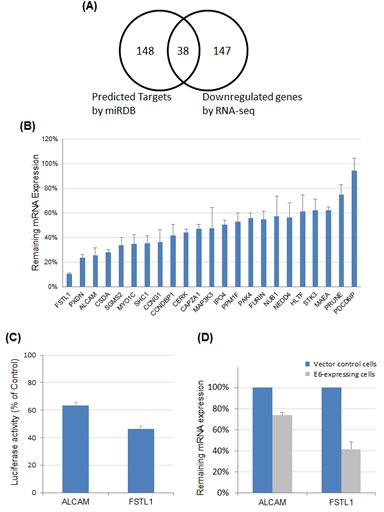
(A) Systematic identification of miR-9 targets by combining computational target prediction with RNA-seq expression profiling. Transcriptome-wide miRNA target prediction was performed with miRDB. In parallel, RNA-seq experiment was performed to globally identify genes downregulated by miR-9 overexpression. Among all genes expressed in HeLa cells, 38 were both predicted miR-9 targets and downregulated by miR-9 as revealed by RNA-seq. (B) Real-time RT-PCR to validate miR-9 targets. Among 23 selected target candidates, 18 were downregulated by over 40% (<60% remaining expression level). (C) Luciferase reporter assays to validate miR-9 target sites in the 3′-UTR of FSTL1 or ALCAM mRNA. The 3′-UTR sequences of FSTL1 and ALCAM were individually cloned into pmirGLO, and luciferase activities were measured after co-transfection of individual 3′-UTR constructs with miR-9 and negative control RNA, respectively. The impact of FSTL1 or ALCAM 3′-UTR on firefly luciferase activity was evaluated by comparing with the negative control and normalized by Renilla luciferase signals. (D) Downregulation of FSTL1 and ALCAM in E6-expressing NIKS cells as compared with vector control cells.
