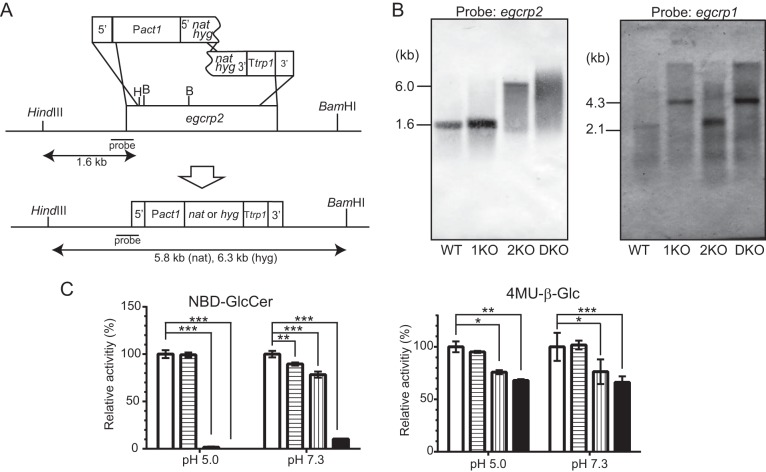
FIGURE 6.
Disruption of the egcrp2 gene in C. neoformans. A, strategy for the disruption of C. neoformans egcrp2 using the split marker method. B, Southern blot analysis of BamHI-HindIII-digested genomic DNA for egcrp2 (left) and HindIII-digested genomic DNA for egcrp1 (right). The disruption of egcrp1 and Southern blot analysis of HindIII-digested genomic DNA were performed by the method described previously (12). C, the activity of β-glucosidase was measured using NBD-GlcCer (left) and 4MU-β-Glc (right). Activities were measured at 37 °C for 18 h using NBD-GlcCer and 2 μg of protein for pH 5.0 or 10 μg of protein for pH 7.3. When 4MU-β-Glc was the substrate, 1 μg of protein was incubated at 37 °C for 18 h for both pH values. The activity was expressed as a percentage of that of WT. *, **, and ***, p < 0.01, p < 0.001, and p < 0.0001, respectively. Error bars, S.D.