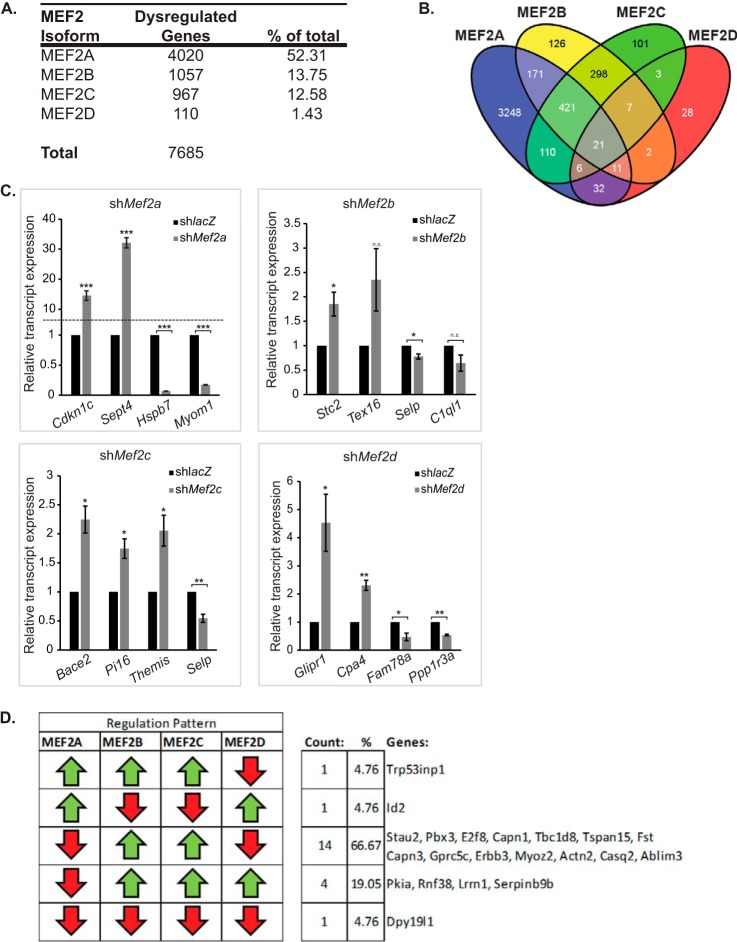
FIGURE 4.
Comparative analysis of MEF2 knockdown gene sets. Microarray analysis reveals that C2C12 cells are differentially sensitive to depletion of the four MEF2 isoforms. A, summary of total significantly dysregulated (q < 0.05) genes in each MEF2 isoform shRNA knockdown. B, a composite Venn diagram incorporating all overlapping gene sets as determined by the Tukey's honest significant difference post hoc test (q < 0.05). C, quantitative RT-PCR analysis of a subset of genes dysregulated in the Mef2 knockdown microarrays. Cdkn1c, cyclin-dependent kinase inhibitor 1c; Sept4, septin 4; Hspb7, heat shock protein family member 7; Myom1, myomesin 1; Stc2, stanniocalcin 2; Tex16, testis expressed gene 16; Selp, selectin, platelet; C1ql1, compliment component 1, q subcomponent-like 1; Bace2, beta-site app-cleaving enzyme 2; Pi16, peptidase inhibitor 16; Themis, thymocyte selection associated; Glipr1, GLI pathogenesis-related 1 (glioma); Cpa4, carboxypeptidase a4; Fam78a, family with sequence similarity 78; Ppp1r3a, protein phosphatase 1, regulatory (inhibitor) subunit 3a. The primer sequences are listed under “Experimental Procedures.” D, only five of the possible patterns of dysregulation are represented in the commonly dysregulated gene set. Of these patterns, the most prevalent group (66%) were genes that were down-regulated in MEF2A or MEF2D depletion and up-regulated in MEF2B or MEF2C depletion. Additionally, only a single gene, Dpy19l1 (DumPY19-like 1), was dysregulated in the same direction upon individual knockdown of each MEF2 factor. The data are means ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.