FIGURE 2.
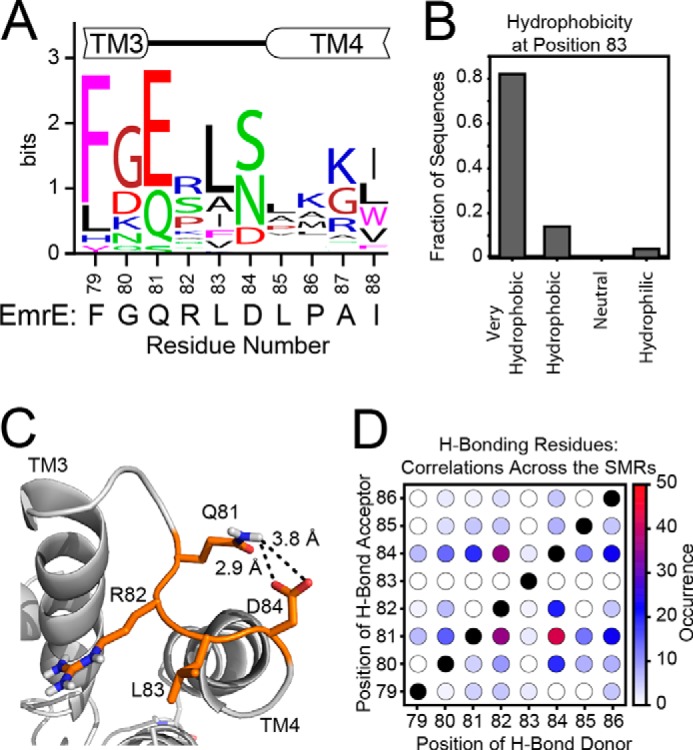
The composition and chemistry of loop-3 is conserved across the SMR family. A, the consensus logo plot for the loop between TM3 and TM4 was generated from the multiple sequence alignment of SMR sequences from the Uniref90 database. The conservation of residue type shows that the loop tends to have a polar or negatively charged residue at position 81, a hydrophobic residue at position 83, and a polar or negatively charged residue at position 84. For comparison, the sequence of EmrE is displayed below the consensus plot. B, a bioinformatics analysis of position 83 reveals that it is occupied by a very hydrophobic residue (Phe, Ile, Trp, Leu, Val, or Met) (49) in more than 80% of sequences analyzed, with another 10% containing a less hydrophobic Ala at this position. C, the 3.8 Å x-ray crystal structure (PDB 3B5D) (7) with the side chains rebuilt shows a possible hydrogen bond between Gln81 and Asp84. D, the occurrences of hydrogen bond acceptors (Asn, Asp, Gln, Glu, His, Ser, Thr, or Tyr) with hydrogen bond donors (Arg, Asn, Gln, His, Lys, Ser, Thr, Trp, or Tyr) were mapped within each of the 50 sequences of the Uniref90 set to determine the possibility of a hydrogen bond within each sequence. The total occurrences across the set were summed and plotted in the heat map. Across the sequences, a hydrogen bond acceptor was frequently found at position 81, and a hydrogen bond donor typically found at position 84.
