FIGURE 3:
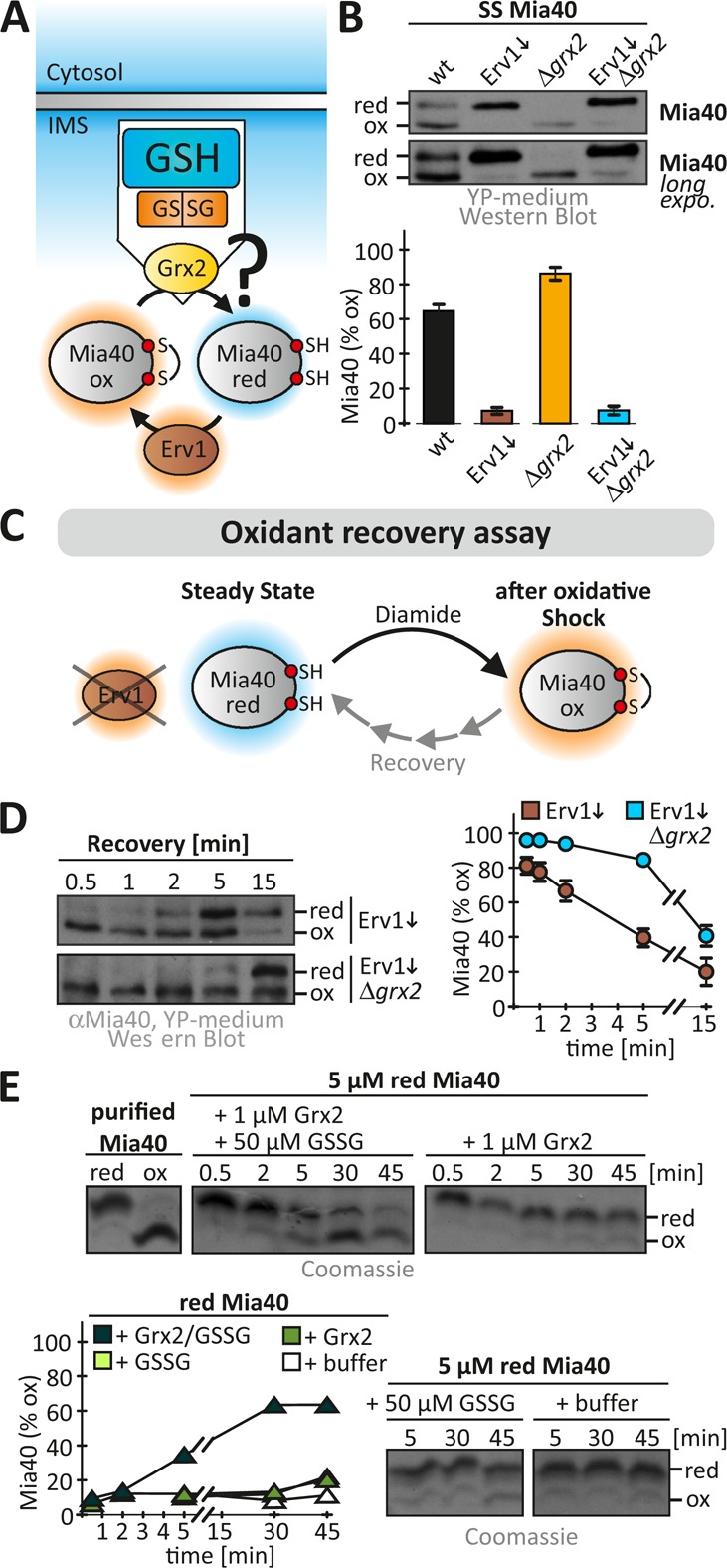
Grx2 directly influences the Mia40 redox state. (A) Scheme depicting redox influences on the IMS oxidoreductase Mia40. Mia40 is oxidized by the sulfhydryl oxidase Erv1 and is reduced by the local glutathione pool potentially mediated by IMS-localized Grxs. (B) Redox state of Mia40 in the indicated yeast cells. Cells grown to mid log phase in YP medium containing glucose to lower Erv1 levels were precipitated with TCA. The resulting pellet was resuspended in a buffer containing 20 mM N-ethylmaleimide (NEM). The samples were analyzed on nonreducing SDS–PAGE, followed by immunoblotting against Mia40. SS, steady state. Reported values are the mean of three independent experiments. Error bars are the means ± SD. (C) Scheme of oxidant recovery assay. Yeast cells were incubated with 20 mM diamide for 5 min, the oxidant was removed, and the recovery of the Mia40 redox state was assessed at different times using an NEM-based alkylation gel shift assay. (D) Oxidant recovery assays in wild-type and Δgrx2 cells, both depleted of Erv1. Reported values are the mean of three independent experiments. Error bars are the means ± SD. (E) In vitro reconstitution of the Grx2-Mia40 disulfide exchange. Reoxidation of purified reduced Mia40 was followed in the presence of GSSG, Grx2, or buffer or a combination of all three. The redox state of Mia40 was assessed at the indicated times using AMS.
