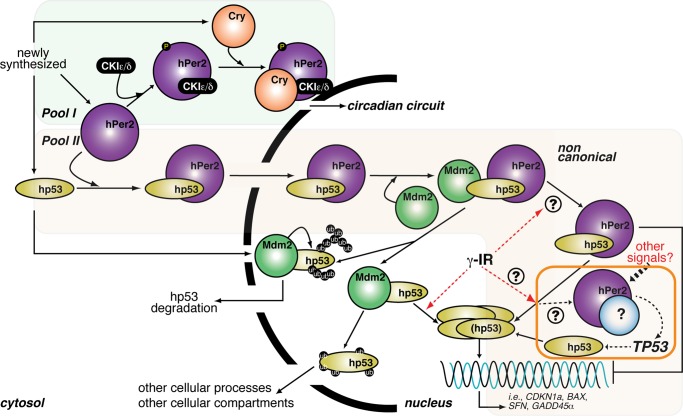
FIGURE 6:
Proposed model of hPer2 and hp53 interaction and function. Newly synthesized hp53, hPer2, and Cry localize in the cytosolic compartment where the Cry/hPer2/CKIε/δ (pool I) is formed, and hPer2 is incorporated in one or more complexes that constitutes pool II. Pool I: As extensively reviewed, cytosolic hPer2 is phosphorylated by casein kinase I ε/δ (CKIε/δ) and initially degraded by the ubiquitin proteasome pathway (Ko and Takahashi, 2006). Later, Cry accumulates and associates with hPer2/CKIε/δ, and this complex translocates to the nucleus, where Cry disrupts the Clock/Bmal1-associated transcriptional complex, resulting in inhibition of CRY, PER, and REB-ERVα and derepression of BMAL1 transcription and modulation of the expression of other clock-controlled genes (shown as “circadian circuit” for simplicity). Pool II: The hPer2 protein associates with cytosolic hp53, forming a stable complex (Gotoh et al., 2014) that translocates to the nuclear compartment and keeps hp53 in a functionally inactive but stable state, ensuring that basal levels of hp53 exist (“priming state”). This heterodimer eventually incorporates Mdm2, forming a trimeric and stable Mdm2/hp53/hPer2 complex. In response to, for example, a genotoxic stress (labeled as γ-IR in the cartoon), the trimeric complex disassembles by a yet-unknown mechanism, which leads to release of hp53 and downstream activation of genes involved in cell cycle arrest and DNA repair. An amplification loop exists in which hPer2, alone or in association with an unidentified partner, transcriptionally activates TP53, further sustaining the hp53-mediated response (boxed in orange). Alternatively, cytosolic hp53 enters the nucleus, where it is targeted by Mdm2 and either polyubiquitinylated and degraded back into the cytosol or monoubiquitinated and translocated to a different compartment, as has been described (for review, see Kruse and Gu, 2009).