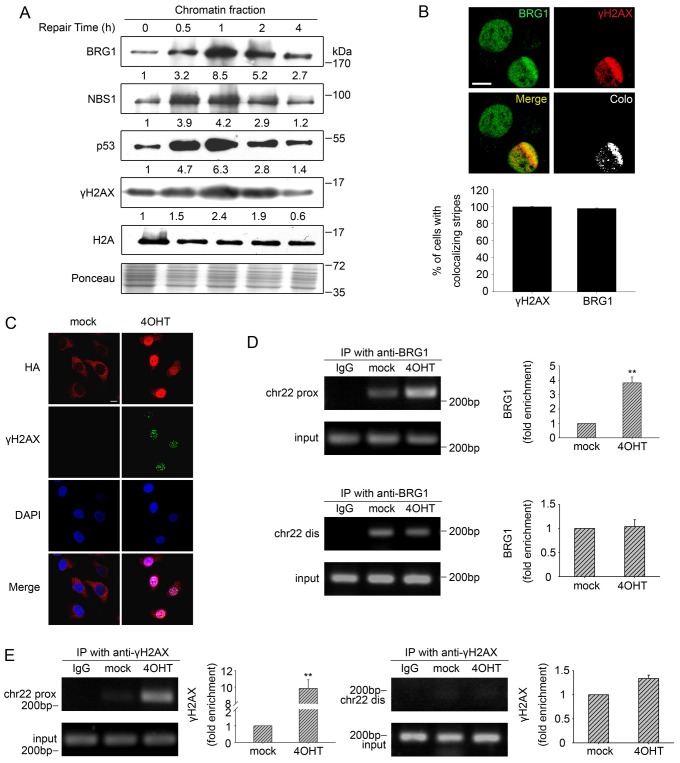
Fig. 2.
BRG1 is enriched on the chromatin at DSBs. (A) U2OS cells were incubated with 10 µM ETO and then allowed to repair DSBs. The chromatin fractions were extracted from cell pellets collected at the indicated time-points and were subjected to immunoblot analysis. Ponceau S staining showed the equal loading of total proteins, and H2A was used to show equal quantities of chromatin fractions from different samples. (B) U2OS cells were subjected to multiphoton laser irradiation. After 1 h, cells were immunostained for endogenous BRG1 and γH2AX. Colocalisation (Colo) of BRG1 and γH2AX in merged images was analysed by using the confocal microscopy software to generate white dots. The percentage of cells with colocalisation of γH2AX with BRG1 is indicated as the mean±s.d. (lower panel). Scale bar: 10 µm. (C) AsiSI–ER-U2OS cells were subjected to 4OHT treatment (3 h) or were mock treated. Then, the cells were fixed and incubated with antibodies against HA–AsiSI and γH2AX. Scale bar: 10 µm. (D) ChIP analysis was performed using AsiSI–ER-U2OS cells with or without 4OHT treatment using an antibody against BRG1 or isotype IgG control antibody. Precipitated DNA was assessed by PCR amplification using primers located in close proximity to (prox) and distal from (dis) the AsiSI site as described in Materials and Methods. IP, immunoprecipitation; chr22, chromosome 22. (E) A ChIP assay was performed using antibody against γH2AX. Quantification of immunoprecipitated material is shown. The values represent the mean±s.d. (three independent experiments); **P<0.01.