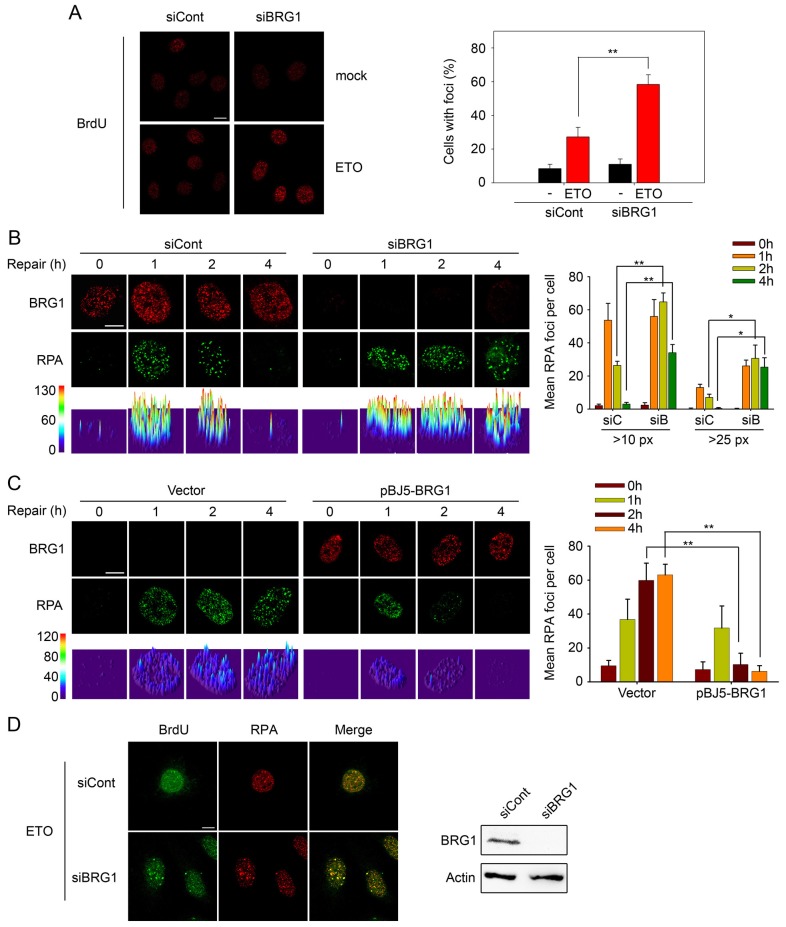
Fig. 4.
Loss of BRG1 results in ssDNA accumulation and retention of RPA foci. (A) U2OS cells transfected with BRG1 siRNA (siBRG1) or control siRNA (siCont) were pre-labelled with BrdU and then treated with 10 µM ETO for 20 min. After 2 h, the cells were fixed under non-denaturing conditions, and BrdU in ssDNA was detected with an anti-BrdU antibody. Representative images are presented. Quantitative analysis of BrdU-positive cells is shown on the right. Cells with more than five foci were considered positive, with an average of 100 cells counted per experiment (three independent experiments). (B) U2OS cells transfected with BRG1 siRNA (siBRG1, siB) or control siRNA (siCont, siC) were exposed to 10 µM ETO for 20 min. At the indicated time-points, cells were pre-extracted with buffer D to release non-chromatin binding proteins, and detected by immunostaining with antibodies recognising BRG1 (red) and RPA (green), respectively. The lower row shows the three-dimensional plot of the intensity of RPA shown in the upper panels, as determined by using Image J software. Quantification of RPA foci [>10 or >25 pixels (px)] is shown on the right. (C) SW13 cells transfected with pBJ5-BRG1 or empty vector were treated and analysed as in B. Foci with >10 pixels were counted. All quantitative data show the mean±s.d.; *P<0.05; **P<0.01. (D) U2OS cells were treated as in A. After 2 h, cells were fixed and detected by immunostaining using anti-BrdU and anti-RPA antibodies. BRG1 expression was analysed by immunoblotting. Scale bars: 10 µm.