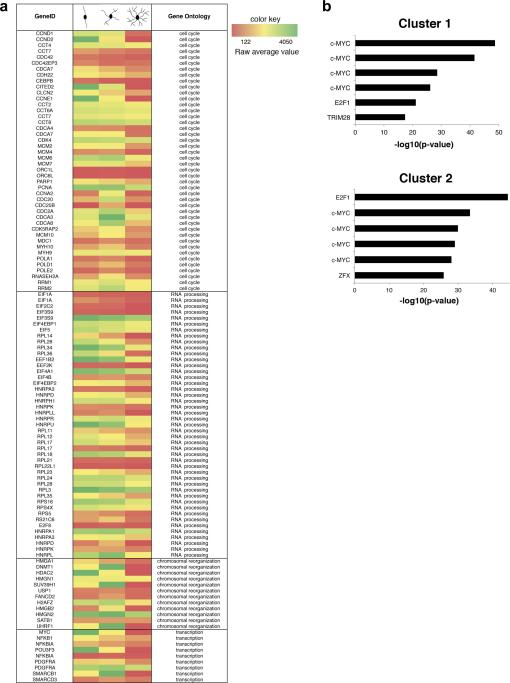
Figure 1. Identification of c-Myc as a relevant transcription factor during oligodendrocyte differentiation.
(a) Heat Map showing the microarray signals of selected transcripts during a defined time course (Dugas et al., 2006) of primary oligodendrocyte progenitor cells (OPCs) differentiation. Only gene ontology categories that were enriched in the first three clusters of co-regulated genes are reported (cell cycle, RNA processing, chromosomal reorganization, transcription). These correspond to early downregulated genes, as previously described (Swiss et al., 2011). (b) ChIP Enrichment Analysis (ChEA) of the co-regulated gene list in clusters 1 and 2. Bar graphs identify the most significant transcription factors with ChIP-validated binding sites. C-Myc is the top ranking transcription factor regulating genes in cluster 1 and the second ranking transcription factor in cluster 2.