Figure 1.
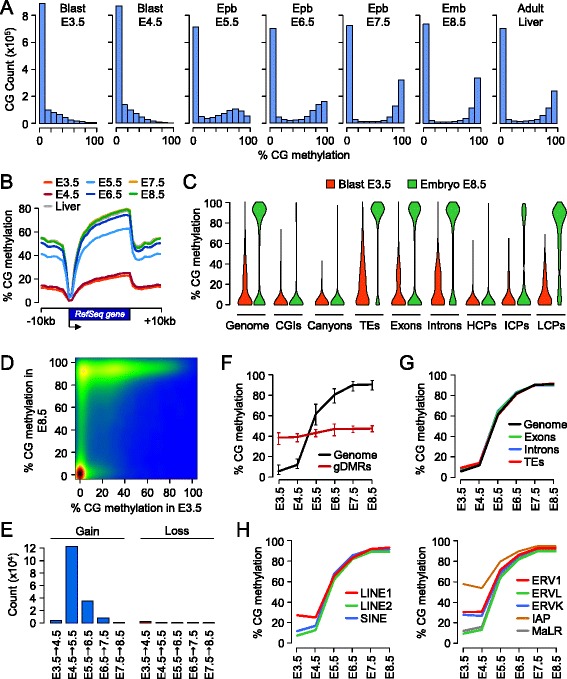
Acquisition of CpG methylation occurs at implantation in mouse embryos. (A) Density histograms showing the distribution of methylation levels measured at individual CpGs throughout embryonic development. Blast, blastocyst; Epb, epiblast; Emb, embryo. (B) Distribution on CpG methylation in RefSeq genes and 10 kb of flanking sequences throughout embryonic development. For each protein-coding RefSeq gene (excluding the X and Y chromosomes), we calculated methylation in 20 equal-sized windows within the gene and 10 1-kb windows of flanking sequences. (C) Violin plots showing the acquisition of CpG methylation in post-implantation embryos compared with blastocysts in various genome elements. TEs, transposable elements; HCPs, high CpG promoters; ICPs, intermediate CpG promoters; LCPs, low CpG promoters. (D) Pairwise comparison of CpG methylation (measured in 400 bp tiles) in E3.5 blastocysts and E8.5 post-implantation embryos. The density of points increases from blue to dark red. (E) Number of 400 bp tiles that gain or lose more than 20% CpG methylation at each developmental transition. (F) Kinetics of de novo DNA methylation in development. We selected all the genomic tiles (400 bp) that gain methylation in post-implantation embryos (defined as <20% methylation in E3.5 blastocysts and >50% methylation in E8.5 embryos) and then plotted their methylation as a function of the developmental stage (black line). The red line shows the methylation for 17 imprinted germline DMRs. The lines represent the median methylation and the error bars represent the 25th and 75th percentiles. (G) Kinetics of de novo DNA methylation in exons, introns and transposable elements (TEs). (H) Kinetics of de novo DNA methylation in classes of transposable elements. LINE, long interspersed nuclear element; SINE, short interspersed nuclear element. In (G,H), the lines depict the median methylation measured at each developmental stage.
