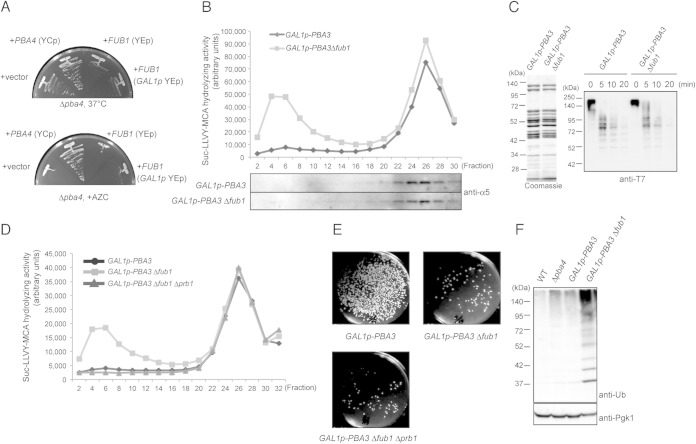
FIG 5.
Fub1 is unlikely to be involved in proteasome assembly. (A) Δpba4 cells were transformed with pYF5 (YEp GAL1p), pT17 (YCp PBA4), pT261 (YEp FUB1), and pT168 (YEp GAL1p-FUB1), and transformed cells were cultured on yeast extract-peptone-galactose (YPGal) for 2 days at 37°C and on YPGal plus 5 mM AZC for 3 days at 27°C. (B) Total lysates of the indicated cells were prepared after the cells were cultured in YPD medium for 2 days to shut off the expression of PBA3 and fractionated by 8 to 32% glycerol gradient centrifugation. Suc-LLVY-MCA hydrolyzing activity was measured in the presence of SDS, and each fraction was subjected to immunoblotting for α5. (C) In vitro degradation of ubiquitinated T7-Sic1PY-6×His. 26S proteasomes were purified from TD132 (GAL1p-PBA3 RPN11-3×Flag) and TD133 (GAL1p-PBA3 Δfub1 RPN11-3×Flag) strains cultured as described for panel B and were subjected to SDS-PAGE and Coomassie blue staining (left). Ubiquitinated T7-Sic1PY-6×His was incubated with the purified 26S proteasomes for the indicated times. Reaction mixtures were subjected to immunoblot analysis using anti-T7 antibodies (right). (D) The Suc-LLVY-MCA hydrolyzing activity of fractionated lysates of the indicated cells was measured in the presence of SDS. (E) Effect of Δprb1 on cell viability. One thousand cells of each indicated genotype were spread on YPGal plates to count the viable cells after PBA3 shutoff for 2 days in YPD medium. (F) Detection of polyubiquitinated proteins in wild-type, YT54 (Δpba4), YT595 (GAL1p-PBA3), and YT745 (GAL1p-PBA3Δfub1) cells cultured as described for panel B. Total lysates of the indicated strains were subjected to SDS-PAGE, followed by immunoblot analysis using antiubiquitin antibodies.