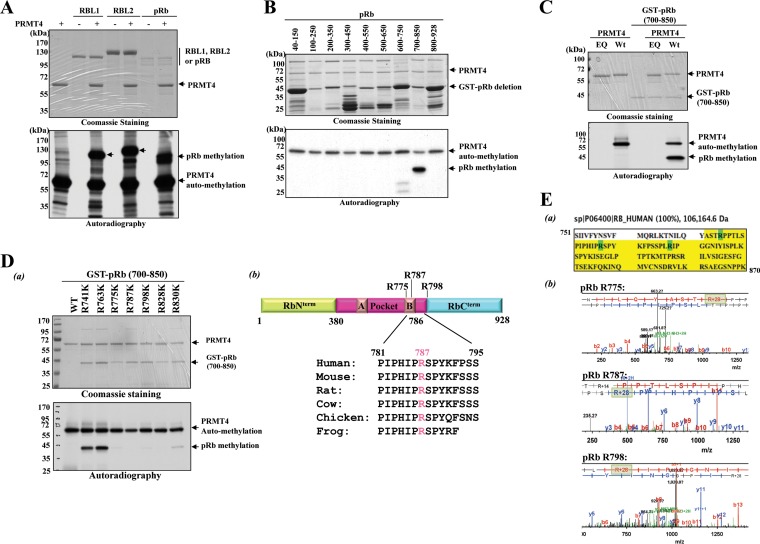
FIG 1.
PRMT4 methylates pRb on the C-terminal domain in vitro. (A) In vitro methylation reactions of purified full-length pRb family proteins (0.5 μg to 2 μg) incubated with S-adenosyl-l-[methyl-3H]methionine as a cosubstrate in the absence or presence of 1 μg PRMT4 (WT). Methylation was detected by autoradiography (bottom), and protein loading was detected by CBB staining after resolution by 8% PAGE (top). (B) (Bottom) Autoradiogram of PRMT4 (WT)-dependent in vitro methylation assay on E. coli-derived overlapping recombinant GST-fused pRb deletion mutants. (Top) CBB staining was carried out as a loading control. (C) In vitro methylation assays with enzymatically inactive PRMT4 (EQ) alone (1st lane), PRMT4 (WT) alone (2nd lane), and the GST-pRb (700–850) fusion protein either alone (3rd lane) or in the presence of PRMT4 (EQ) (4th lane) or PRMT4 (WT) (5th lane). Reactions were stopped in SDS sample buffer, and proteins were separated by 10% PAGE, followed by autoradiography for the detection of methylation (bottom) and CBB staining for a loading control (top). (D) (a) Arginine (R)-to-lysine (K) point mutations were introduced for each R residue within the GST-pRb (700–850) fusion protein, and the resulting point mutants were subjected to in vitro methylation assays. Methylation was detected by autoradiography (bottom) after separation by 10% SDS-PAGE. CBB staining was applied for a protein loading control (top). (b) (Top) Schematic representation of full-length pRb and the relative positioning of the arginine methylation sites. (Bottom) Conservation of the R787 amino acid residue in various species as indicated. (E) (a) The GST-pRb (751–870) fusion protein was subjected to an in vitro methylation assay and was resolved by 10% SDS-PAGE. After CBB staining, the band corresponding to the GST-pRb fusion protein was cut out and was analyzed by liquid chromatography-mass spectrometry at the UC Davis Proteomics Core Facility. The protein sequence covered in the analysis is highlighted in yellow. Dimethyl-modified residues are highlighted in green. (b) In vitro methylation reaction of the pRb (700–850) fusion protein, PRMT4 (WT), and cold SAM followed by LC-MS-MS analysis. Only the spectra from digested peptides harboring the methylated R residues identified are included in this figure. Increases of 28 Da in the mass-to-charge ratio are highlighted in light green.