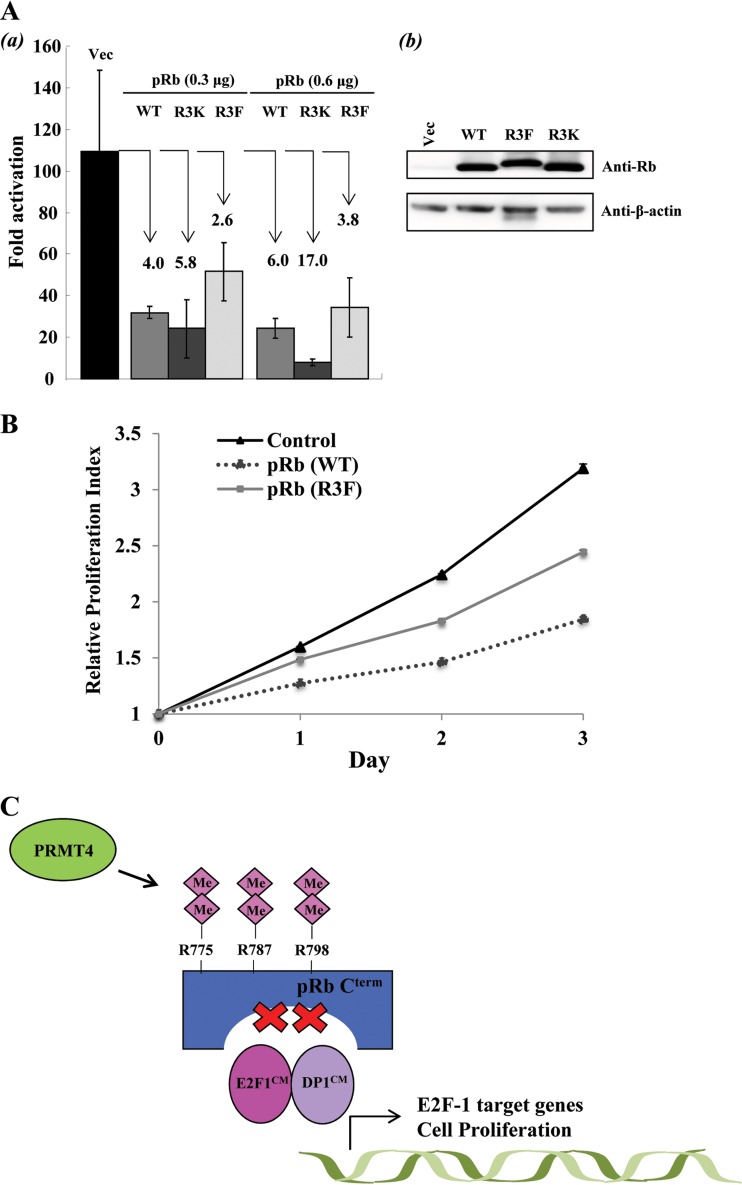
FIG 6.
Arginine methylation inhibits the tumor suppressor function of pRb. (A) (a) Luciferase reporter assays were employed to measure E2F-1 transcriptional activation by cotransfecting HEK293T cells with increasing amounts of DNA plasmids (0.3 μg and 0.6 μg) encoding Flag-pRb (WT), Flag-pRb (R3K), or 3×Flag-pRb (R3F) with expression plasmids encoding the GAL4 DNA binding domain fused to the E2F-1 transactivation domain (residue 190 to the C terminus) and a GAL4 DNA binding site promoter luciferase reporter plasmid. Luciferase activity was measured in arbitrary units by using a Lumat LB 9501 luminometer. The data are averages for triplicate wells. Error bars represent standard deviations (n = 3). The degree of repression is shown for each Rb construct. (b) Immunoblot analysis showing transient expression levels of pRb (WT) and mutants. (B) Cell proliferation studies were performed on U2OS control cells and isogenic U2OS cells expressing either pRb (WT) or pRb (R3F) by using the MTT colorimetric assay. Error bars represent standard deviations (n = 3). The relative proliferation index for each day was determined by measuring the optical density of the cells at 570 nm with a reference filter of 630 nm and using the absorbance from day zero as a standard. (C) Possible model for PRMT4-induced cell cycle progression. During the G1-to-S-phase transition, PRMT4-mediated arginine dimethylation of pRb may facilitate efficient Cdk-dependent phosphorylation of pRb Cterm. Arginine dimethylation also inactivates the tumor suppressor activity of pRb independently of phosphorylation, resulting in E2F-1 dissociation and activation of E2F-1 cell cycle target genes. Me, methylation.