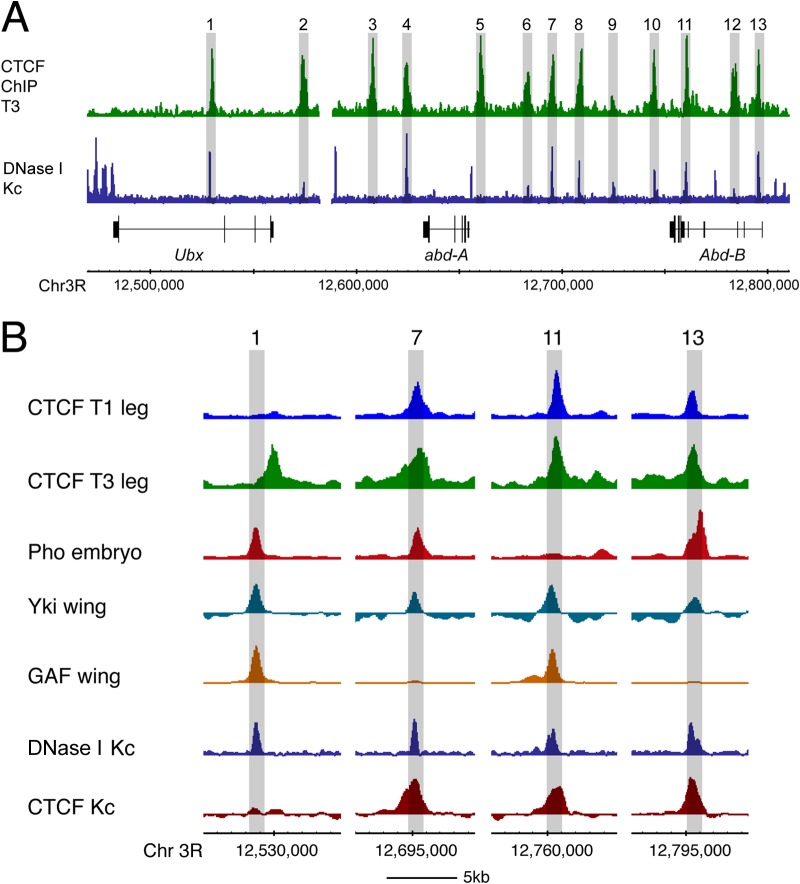
FIG 7.
Chromatin accessibility and protein binding at CTCF sites in the BX-C. (A) In the repressed BX-C in the Drosophila Kc167 cell line (Kc), DNase I profiling reveals specific accessible sites in the repressed chromatin. Thirteen CTCF sites, bound in T3 chromatin, are numbered; 11 of the 13 are associated with DNase I sensitivity peaks. (B) Close-up of selected sites from the experiment whose results are shown in panel A; the binding peaks of several regulators align with the DNase I sites. The variable CTCF site (site 1) is offset from this alignment, whereas other, constitutive CTCF sites are more closely aligned with the DNase I sites. Data from the CTCF T1 and T3 leg are from this paper; data for Pho are from reference 64; data for Yki and GAF are from reference 63; data for DNase I Kc167 are from reference 65; and data for CTCF Kc167 are from ModENCODE (www.modencode.org) data set 908.