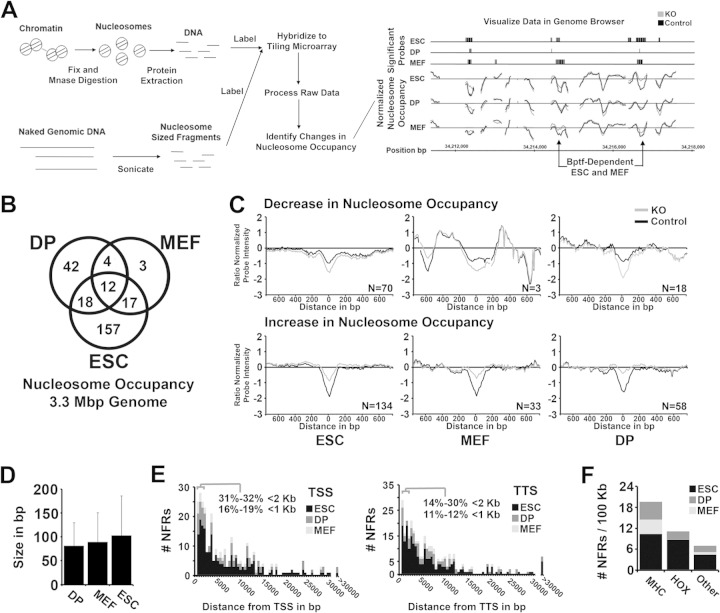
FIG 2.
Bptf regulates nucleosome occupancy at nucleosome-free regions (NFRs) in diverse cell types. (A) Cartoon showing work flow for the digestion, extraction, hybridization, and analysis and visualization of significant changes in nucleosome occupancy with Bptf KO in ESCs, MEFs, and DP thymocytes (see Materials and Methods for detailed procedures). (B) Venn diagram analysis of Bptf-dependent changes in nucleosome occupancy identified from ESCs, MEFs, and DP thymocytes (P values of overlap by Fisher's exact test: DP+MEF, 1.7E−13; ESC+MEF, 3.1E−24; ESC+DP, 8.9E−13). (C) Aggregate plots showing Bptf-dependent changes in nucleosome occupancy and their position at the NFR. Nucleosome occupancy is measured on the y axis as a Z-score-normalized log2 nucleosomal-DNA/genomic-DNA signal ratio from our tiling array (average from 3 biological replicates). Zero on the y axis represents the average nucleosome occupancy across the genome sampled by our tiling array. Individual traces were centered on the x axis at the probe with the greatest change in nucleosome occupancy with Bptf KO. Traces are aggregated based on an increase or decrease in nucleosome occupancy at the NFR with Bptf KO and by cell type. The number of traces used to make each aggregate is included inside each plot. (D) Bar graph showing average size of a Bptf-dependent change in the NFR from ESCs, MEFs, and DP thymocytes. The size of a change in NFR is measured by the number of consecutive tiled probes (60-bp probes tiled at 10-bp intervals) which significantly change in hybridization signal with Bptf KO. (E) Histogram plot showing the distance of a Bptf-dependent NFR to the closest transcription start site (TSS) or closest transcription termination site (TTS) from reference sequence (Ref Seq) genes. (F) Density of Bptf-dependent NFRs (NFRs per 100 kb) at the MHC and HOX genes (A, B, and C clusters) and all other genes analyzed. A higher density of Bptf-dependent NFRs is observed at the MHC and HOX genes than across the whole genome.