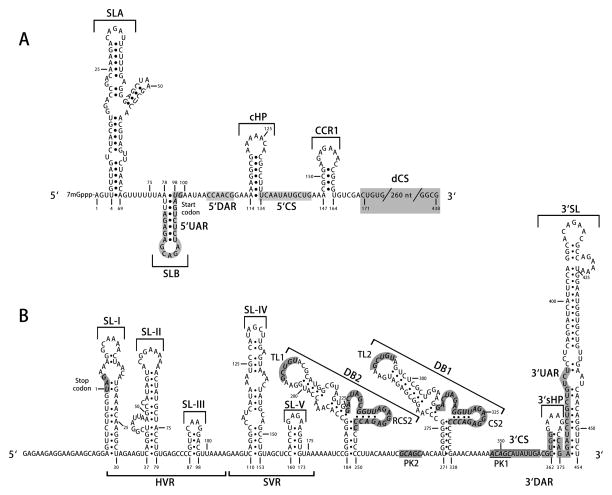
Figure 3. Schematic representation of conserved sequences and structures in the DENV untranslated regions (UTRs) and capsid-coding region.
(A) 5’UTR and capsid-coding region. The DENV 5’UTR contains one large stem-loop (SLA) and one short stem-loop (SLB). The 5’ upstream AUG region (5’UAR) is located within SLB. Downstream of the start codon, within the capsid-coding region, there are several cis-acting RNA elements, including a short 5’ downstream of AUG region (5’DAR), capsid-coding region hairpin (cHP), 5’ cyclization sequences (5’CS), conserved capsid-coding region 1 (CCR1), and downstream of the DENV 5’ cyclization sequence (dCS) element. (B) 3’UTR. The 3’UTR begins with a variable region (VR) containing the hypervariable region (HVR) and semi-variable region (SVR), followed by two dumbbell structures in tandem (DB2 and DB1), 3’ cyclization sequences (3’CS), 3’ downstream of AUG region (3’DAR), and 3’ upstream AUG region (3’UAR). The latter two RNA structures overlap with the 3’ stem-loop (3’SL), which includes a small 3’-end hairpin structure (3’sHP) and a large stem-loop. The top loops (TLs) of the dumbbells are proposed to interact with downstream pseudoknot (PK) regions. The 3’UTR nucleotides are numbered starting from and including the stop codon. Nucleotide numbering is according to DENV2 strain 16681.