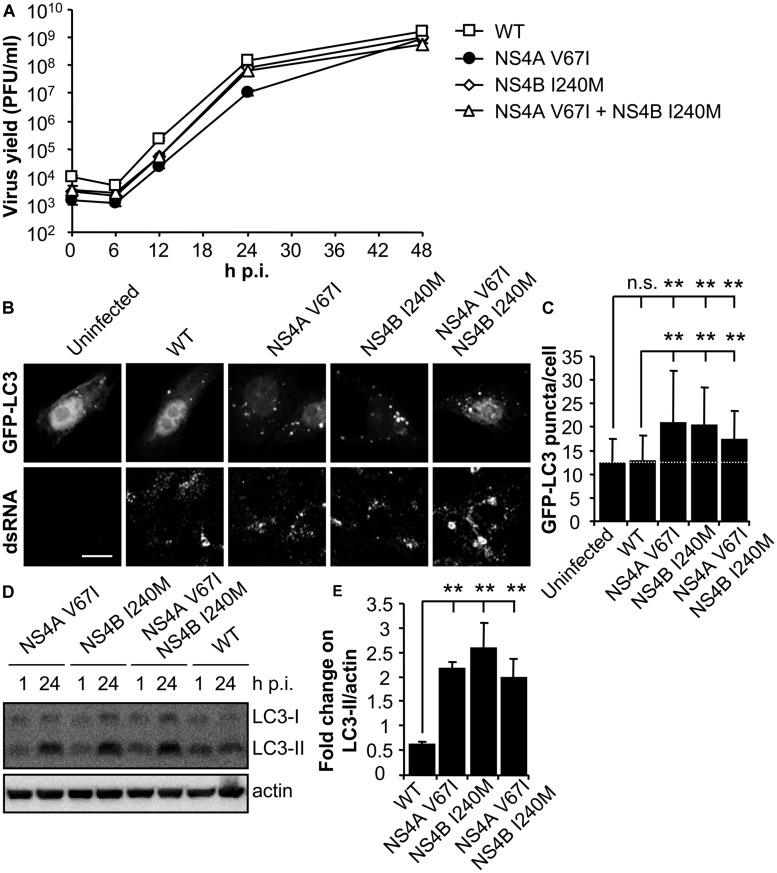
FIGURE 3.
Mutations on WNV NS4A or NS4B can modulate the induction of LC3 modification and aggregation in cells infected with WNV. (A) Growth curves of recombinant WNV recovered from infectious cDNA clone pFLWNV (WT) or its derivatives carrying amino acid substitutions NS4A V67I, NS4B I240M or NS4A V67I + NS4B I240M. Vero cells were infected (MOI of 0.5 PFU/cell) with the viruses and the supernatant virus yield was determined at different times p.i. (B) Visualization of autophagosome formation by LC3 aggregation in cells infected with the viruses displayed in panel (A). Vero cells were transfected with a plasmid encoding GFP-LC3 and 24 h post-transfection were infected (MOI of 5 PFU/cell), fixed and processed for immunofluorescence analysis as described in the legend for Figure 1. Scale bars: 10 μm. (C) The number of fluorescent aggregates on the cytoplasm of cells infected in (B) was determined. Dashed line indicates the mean number of GFP puncta aggregates found in uninfected cells. (D) Analysis of LC3 modification following infection with recombinant WNVs. Vero cells were infected (MOI of 0.5 PFU/cell) and lysed at 1 or 24 h p.i. Western blot analysis was performed as described in the legend of Figure 1. (E) Quantification of the fold change on LC3-II/actin from 1 to 24 h p.i. Results are product of three independent western blots similar to that displayed in panel (D). Statistically significant differences between infected and uninfected cells, or cells infected with WT and mutant viruses, are denoted by two asterisks (**) for P < 0.005. n.s. denotes not statistically significant differences.