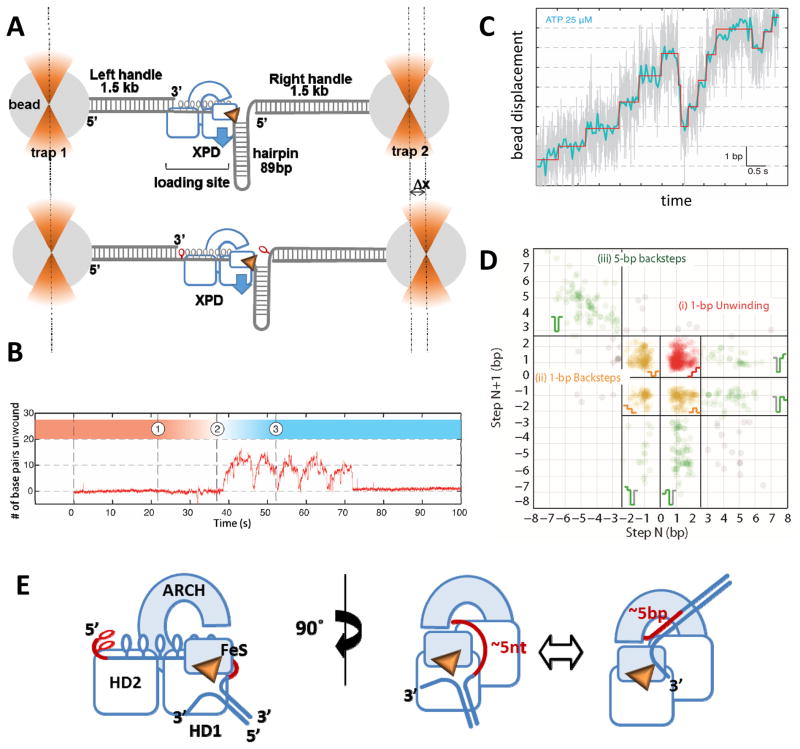
Figure 4. Analysis of XPD stepping kinetics in the high-resolution optical tweezers experiment.
A. The construct for monitoring XPD helicase activity consisted of a dumbbell DNA structure, in which two 1.5 kb handles were tethered to the polystyrene beads held in the optical traps, and an 89 bp hairpin structure flacked by a ssDNA region that served as a loading site for XPD. Blue arrow depicts direction of the XPD movement along the hairpin. Each unwound base pair lengthens the construct by 2 nucleotides (shown in red). The substrate is constructed in a way that allows only the hairpin to be unwound and not the handles (see [67] for details). B. A representative unwinding trajectory. After dumbbell incubation in the XPD containing channel (1) the construct is transferred into the ATP-containing channel (2) where unwinding is detected under the constant applied force as the change in the length of the dumbbell structure resulting in the change in the position of the right bead. This particular trajectory shows 5 consecutive attempts at hairpin unwinding by a single XPD molecule. Each unwinding burst is followed by sliding of the helicase back to the hairpin base. C. A fragment of the representative unwinding trajectory with actual extension data (grey) overlaid with filtered data (blue) and steps determined from fitting the data (red). D. Representative scatter plot of step pairs. Each data points represent the size of every adjacent pair of steps. Consecutive forward steps at highlighted in red, the pairs consisting of at least one backward step are in orange, and 5-bp steps are in green. The data shown were adapted from [67]. E. Proposed mechanics of 5-bp stepping.