Figure 2.
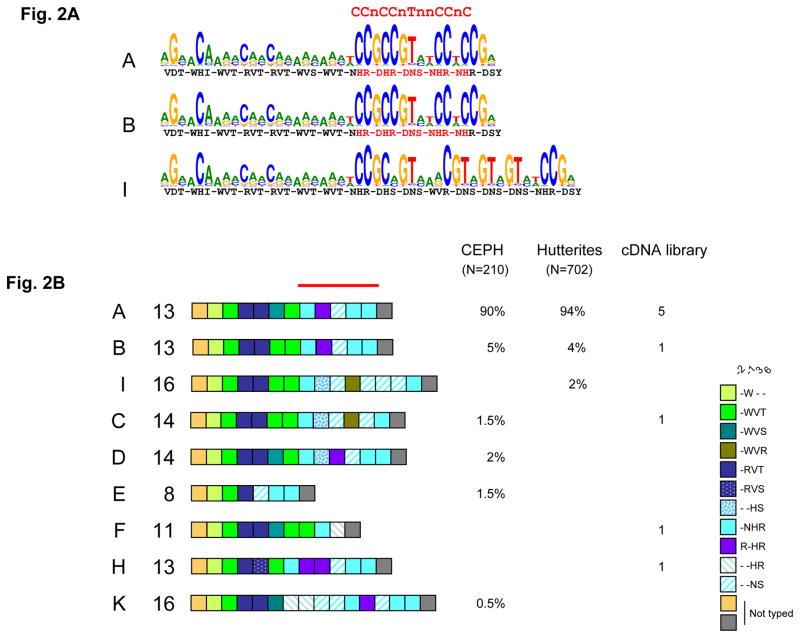
A. Human PRDM9 major alleles (A and B) are predicted to bind the 13-mer hotspot motif, whereas the I allele is predicted to bind a distinct motif. The LD-based hotspot consensus identified by Myers et al. (13) is shown above. The amino-acids at position −1, 3 and 6 of the zinc finger alpha helices are indicated as in figure 1B, with the residues predicted to recognize the LD-hotspot consensus motif shown in red.
B. Allelic diversity of the human PRDM9 zinc finger tandem array.
Interspersion patterns of variant repeats (colored boxes) of alleles from unrelated individuals was established either by Minisatellite Variant Repeat mapping (105 CEPH unrelated parents or grand-parents and 351 Hutterite parents) or by sequencing clones from a testis cDNA library made from 39 donors. Major allele A and minor allele B were found in all three sets of unrelated individuals and other rare alleles only in one or two sets. The structures of some rare alleles (I, C, E and F) differ strongly from A and B in the region encoding the critical domain (red bar) for recognition of the 13-mer hotspot motif.